Published online Oct 28, 2011. doi: 10.3748/wjg.v17.i40.4496
Revised: June 2, 2011
Accepted: June 9, 2011
Published online: October 28, 2011
AIM: To elucidate cell proliferation in erosive reflux disease (ERD) and non-erosive reflux disease (NERD), we evaluated markers in squamous epithelial cells.
METHODS: Thirty-four consecutive patients with gastroesophageal-reflux-disease-related symptoms (21 NERD and 13 ERD) were evaluated for the enrolment into the study. All patients underwent 24-h pH monitoring, standard endoscopy, and biopsy for histological evaluation. The expression of cyclins D and A was evaluated by real-time reverse transcription polymerase chain reaction (RT-PCR) from isolated epithelial cells. In all samples, analysis of the isolated cell population revealed the presence of epithelial cells only.
RESULTS: Real-time RT-PCR showed that, in patients with ERD, the relative expression of cyclin D1 mRNA in esophageal epithelium was strongly decreased in comparison with NERD patients. The mean value of relative expression of cyclin D1 mRNA in NERD patients was 3.44 ± 1.9, whereas in ERD patients, it was 1.32 ± 0.87 (P = 0.011). Real-time RT-PCR showed that, in patients with ERD, relative expression of cyclin A mRNA in esophageal epithelium was decreased in comparison with that in NERD patients (2.31 ± 2.87 vs 0.66 ± 1.11). The mean bromodeoxyuridine labeling index in the NERD patients was 5.42% ± 1.68%, whereas in ERD patients, it was 4.3% ± 1.59%.
CONCLUSION: We confirmed reduced epithelial proliferation in ERD compared with NERD patients, and that individuals who develop ERD are characterized by weaker epithelial cell proliferation.
- Citation: Calabrese C, Montanaro L, Liguori G, Brighenti E, Vici M, Gionchetti P, Rizzello F, Campieri M, Derenzini M, Trerè D. Cell proliferation of esophageal squamous epithelium in erosive and non-erosive reflux disease. World J Gastroenterol 2011; 17(40): 4496-4502
- URL: https://www.wjgnet.com/1007-9327/full/v17/i40/4496.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i40.4496
Most of the patients with gastroesophageal reflux disease (GERD) fall into one of two categories: non-erosive reflux disease (NERD) or erosive reflux disease (ERD). The two main phenotypes of GERD appear to have different pathophysiological and clinical characteristics. NERD is the most common phenotypic presentation of GERD. Although separation of ERD and NERD on a clinical level is difficult, there are clearly physiological, pathophysiological, anatomical, and even histological characteristics that are unique to NERD. Natural course studies have demonstrated that most NERD patients do not progress over time to ERD or even Barrett’s esophagus. NERD patients compared to those with ERD demonstrate a highly variable and unpredictable symptomatic response rate to antireflux treatment[1].
Cell replication of basal layers is hypothesized to be one of the causes implicated in the resistance of the mucosa and structural epithelial defense. In previous investigations, we have demonstrated that, in patients with GERD, the number of proliferating cells, evaluated by Ki-67 immunostaining, was reduced in esophageal mucosa exposed to chronic acid-peptic insult[2,3]. Two reasonable hypotheses can be suggested to explain the reduced epithelial proliferation activity observed in GERD: (1) chronic cell damage induced by GER determines a reduction in the proliferation rate of esophageal epithelium; or (2) a constitutive lower capacity for cell proliferation brings a major susceptibility to damage induced by GER.
Our findings are in contrast to the results of a recent study[4] on the cell proliferation of squamous epithelium in GERD. This study has shown a significantly higher number of proliferating cells in GERD patients compared with that in controls, as evaluated by Ki-67 immunostaining.
To elucidate the different proliferation in NERD and ERD patients, the present study evaluated squamous epithelial cell proliferation in patients with GERD, in comparison with NERD, by measuring the S-phase fraction using the bromodeoxyuridine labeling index (BrdU-LI), and by quantifying the expression of cyclins A and D, which are associated with cell cycle progression.
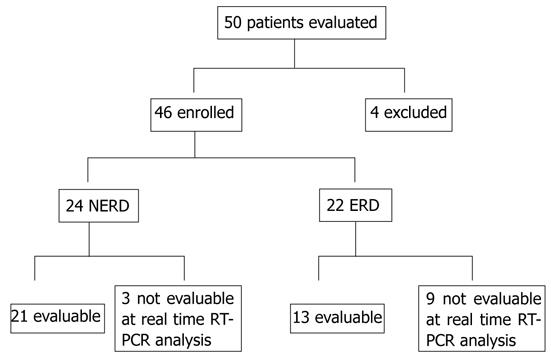
Fifty consecutive patients with GERD-related symptoms were evaluated for enrolment into the study. Inclusion criteria were the presence of typical symptoms (heartburn and⁄or regurgitation) for at least 1 year (frequency was > 2 times/wk) and abnormal 24-h pH parameters and symptom-association probability (SAP). Exclusion criteria were patients with esophageal or gastric malignancy or histologically proven Barrett’s esophagus, gastric or duodenal ulcer, previous esophageal or gastric surgery, extra-esophageal symptoms, patients taking antisecretory or prokinetic drugs at least 30 and 15 d before the procedure, respectively. Forty-six patients (mean age 45.2 ± 13.4 years, range 22-78 years; 20 men) fulfilled the inclusion⁄exclusion criteria and were evaluated. All these patients underwent standard endoscopy and biopsy for histological evaluation. Twenty-four had an apparently normal esophageal mucosa at endoscopy (NERD), whereas 22 had ERD. None of the patients had received cyclical therapy with proton pump inhibitors (PPIs) (not more than 8 wk in the past year). This study was single-blinded for the pH, histological, immunostaining and real-time reverse transcription polymerase chain reaction (RT-PCR) evaluations.
The frequency and intensity of symptoms and their impact on quality of life were registered using a structured and validated questionnaire for the diagnosis of GERD[5], and patients with a score > 3.1 were considered positive. For real-time RT-PCR, only 34 patients (mean age 47.08 ± 16.04 years, range 22-73 years; 16 men) were evaluable (Figure 1). Twenty-one had an apparently normal esophageal mucosa at endoscopy (NERD), whereas 13 had ERD (Table 1).
| NERD | ERD | |
| No. of subjects | 21 | 13 |
| Sex (M/F) | 9/12 | 7/6 |
| Mean age ± SD (yr) (range) | 44.2 ± 14.9 (22-73) | 54.4 ± 15.7 (29-78) |
| Endoscopy | ||
| Normal | 21 | 0 |
| A | 0 | 0 |
| B | 0 | 7 |
| C | 0 | 6 |
| D | 0 | 0 |
| Histology | ||
| Normal | 21 | 12 |
| Mild | 0 | 1 |
| Moderate | 0 | 0 |
| Severe | 0 | 0 |
| 24-h pH monitoring | ||
| Mean % of acid exposure time ± SD | 10.4 ± 1.3 | 10.7 ± 1.4 |
| Mean number of acid reflux events ± SD | 126 ± 20 | 128 ± 22 |
Patients gave written informed consent to participate in the study, which was approved by the local research ethical committee.
Every patient underwent 24-h esophageal pH monitoring according to standard methodology. To define better the localization of the lower esophageal sphincter (LES) and upper esophageal sphincter (UES), esophageal manometry was performed before pH monitoring, with a water-perfused catheter that incorporated three distal openings, radially oriented for LES pressure recording, and three side-hole recording sites at 5, 10 and 15 cm above the distal openings. Multichannel 24-h pH monitoring was performed using two probes, with one and two antimony sensors, respectively, with a separate skin reference (Zinetics Medical Inc., Salt Lake City, UT, United States). In accordance with manometric findings, the three pH sensors were placed at the gastric level, at 5 cm above the LES and 10 cm below the UES, respectively. Data were stored on a single portable digital recorder (Digitrapper pH 200; Medtronic, Minneapolis, MN, United States). Before each study, the pH probe was calibrated in buffer solutions of pH 7 and pH 1.
During the test day, meal time and composition were standardized. The reflux parameters were assessed according to Johnson and DeMeester[6]. Of these, only the percentage of time spent at pH < 4.0 over 24 h was evaluated. The pH testing was considered abnormal if pH < 4.0 was present for > 5% of the total 24 h. The SAP was calculated according to Weusten et al[7] and was considered positive if it exceeded 95%.
Patients underwent upper gastrointestinal (GI) endoscopy (videogastroscope Olympus GIF 160) after sedation by intravenous midazolam (2.5 mg), to assess the presence or absence of erosions.
The Los Angeles classification was used to grade esophagitis[8]. In each subject, eight specimens were taken with standardized biopsy forceps (Olympus FB 24K), from each of the four quadrants, two bites from each quadrant, 5 cm above the squamous-columnar junction (SCJ), from macroscopically intact (non-eroded) esophageal mucosa. The SCJ (or Z-line) was defined as the border between gastric glandular and esophageal squamous epithelium, and it roughly corresponded to the proximal edge of the gastric folds.
Of the eight biopsies taken during endoscopy from each patient, two were used for total RNA extraction, and two for BrdU labeling. Four were oriented to appropriate cellulose acetate supports (Endofilters Bioptica, Milan, Italy), fixed in 4% buffered formalin, and embedded in paraffin, for processing by hematoxylin-eosin for histological and immunohistochemical analysis.
Four-micrometer-thick serial sections were cut from each paraffin block and stained with hematoxylin-eosin. For each case, whole longitudinally sectioned samples were examined. Esophagitis was identified and graded according to the classification of Ismail-Beigi et al[9]: (1) the degree of basal cell hyperplasia, expressed as a percentage of epithelial thickness: none (0%-15%), mild (16%-33%), moderate (34%-67%), severe (> 67%); (2) presence or absence of papillary zone elongation, determined by calculating papillary length as a percentage of epithelial thickness: absent (0%-67%) and present (> 67%); and (3) density of neutrophil and eosinophil infiltration: none (0/high power field), mild (1-2/high power field), moderate 3-10⁄high power field) and severe (> 10/high power field). The area of one high power field was 0.229 mm.
Each biopsy was immersed in 5 mL RPMI 1640 containing non-essential amino acids, 100 U/mL penicillin, 100 μg/mL streptomycin and 10% fetal bovine serum (FBS) supplemented with 160 μmol/L BrdU (Sigma-Aldrich, St Louis, MO, United States) and incubated for 4 h at 37 °C in a 5% CO2/air incubator. Tissues were rapidly rinsed with three washes of cold phosphate buffered saline (PBS), fixed in 10% buffered formalin, and embedded in paraffin. Sections were cut from each paraffin block and picked up on poly-L-lysine-coated slides. Sections were dewaxed, hydrated through decreasing concentrations of ethanol, rinsed in distilled water, and autoclaved in 10 mmol/L sodium citrate buffer (pH 6.0) at 120 °C for 21 min for antigen retrieval. After cooling and washing, the endogenous peroxidase activity was quenched using 3% hydrogen peroxide in absolute methanol for 10 min at room temperature. Sections were incubated with primary mouse anti-BrdU antibody (Bu20a; Abcam, Cambridge, United Kingdom) diluted in 1% bovine serum albumin in PBS overnight at 4 °C, using appropriate dilutions.
Sections were processed according to a non-biotin amplified method (NovoLinkTM Polymer Detection System; Novocastra Laboratories, Newcastle Upon Tyne, United Kingdom) and counterstained with hematoxylin.
Quantitative analysis of BrdU immunostaining was performed on contiguous field visualized on the color monitor of a personal computer equipped with a 3 CCD (charge-couple device) color video camera (KY F55B; JVC, Pinebrook, NJ, United States) connected to a light microscope (Leitz DIAPLAN, Wetzlar, Germany). For each case, whole longitudinally sectioned samples were examined. Samples that did not contain at least 1000 cells were excluded. Quantitative evaluation was only carried out on portions of epithelium in between vertically sectioned stromal papillae, and corresponding to 100 μm from the basal layer. BrdU-LI was defined as the ratio of BrdU-positive nuclei to the total number of epithelial cells, and was expressed as a percentage.
Total RNA was extracted from esophageal epithelial cells that were isolated as follows. Esophageal biopsy samples were washed in PBS and incubated in 0.5% collagenase type II (C6885; Sigma-Aldrich) in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer for 30 min at 37 °C in a shaking bath.
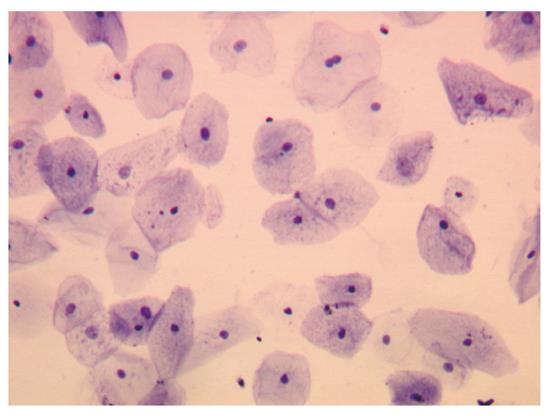
Collagenase activity was blocked by adding the same volume of 20% FBS in HEPES buffer. The digested material was re-suspended and passed through a 40-μm pore size cell strainer (BD Falcon™, Franklin Lakes, NJ United States) and centrifuged for 5 min at 800 g. Cells were washed in PBS and counted in a hemocytometer. From each biopsy sample, an average of 50 000 cells were recovered. Cell morphology was evaluated by seeding cells on poly-L-lysine-coated slides for 2 h at 37 °C. Cells were fixed with 2% paraformaldehyde in PBS for 5 min and stained with 1% toluidine blue in distilled water for 1 min (Figure 2).
Total RNA was extracted from isolated esophageal epithelial cells using TRI Reagent (Ambion, Austin, TX, United States) according to the manufacturer’s instructions. Whole cell RNA was quantified spectrophotometrically and 2 μg RNA for each sample was reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, United States), following the manufacturer’s protocol.
The relative expression of cyclin A (CCNA1), cyclin D1 (CCND1) and the housekeeping gene β-glucuronidase were evaluated by real-time polymerase chain reaction (PCR) performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using TaqMan Gene Expression Assay primers and probe kits (Assays catalog number Hs00927505 for CCNA1 and Hs00277039 for CCND1; Applied Biosystems). Cycling conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. For each sample, three replicates were analyzed. The relative amounts of the transcripts were calculated with the 2-ΔΔCT method against aliquots from a single preparation of calibrator cDNA from the U2OS cell line.
Differences between groups were assessed by Student’s t test. P < 0.05 was considered statistically significant. Data were analyzed with SPSS software (SPSS, Chicago, IL, United States).
At pH monitoring, the percentage time with esophageal pH < 4 in the two groups of patients (NERD and ERD) was 10.4% ± 1.3% and 10.7% ± 1.4%, respectively. No significant differences were found in the mean percentage time between the two groups.
Histological analysis showed that, among 13 patients affected by erosive esophagitis in endoscopic normal mucosa, 12 had a normal pattern, and 1 had mild esophagitis. None of the patients with NERD showed signs of esophagitis (Table 1).
Expression of cyclins A and D was evaluated by real-time RT-PCR from isolated epithelial cells. To check for purity of isolated cells, morphology was assessed after toluidine blue staining (Figure 2). In all samples evaluated, analysis of the isolated cell population revealed the presence of epithelial cells only.
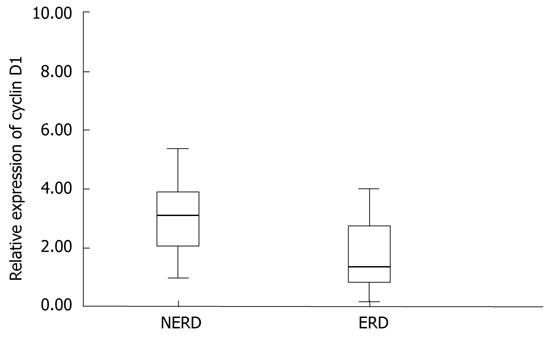
Real-time RT-PCR analysis shows that, in patients with ERD, the relative expression of cyclin D1 mRNA, in esophageal epithelium, was strongly decreased in comparison with that of NERD patients (Figure 3). In particular, the relative expression of cyclin D1 mRNA in NERD epithelium was twofold higher, and showed elevated variability between patients, with respect to ERD epithelium. The relative expression of cyclin D1 mRNA ranged from 0.17 to 8.36 among all patients, with a mean (± SD) value of 2.41 ± 1.8. The mean (± SD) cyclin D1 value in 21 NERD patients was 3.44 ± 1.9, whereas in 13 ERD patients, it was 1.32 ± 0.87 (P = 0.011).
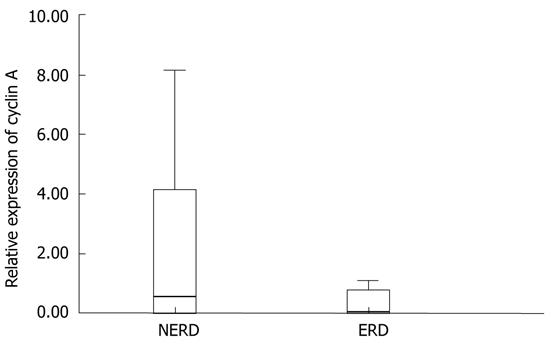
Only 25 of the 34 patients enrolled were evaluable for real-time RT-PCR analysis of cyclin A mRNA (18 NERD and 7 ERD). The relative expression of cyclin A mRNA ranged from 0 to 8.13 among all patients with a mean (± SD) value of 1.84 ± 2.59. Real-time RT-PCR analysis showed that, in patients with ERD, the relative expression of cyclin A mRNA in esophageal epithelium was decreased in comparison with that in NERD patients (Figure 4). In particular, the mean (± SD) cyclin A value of NERD patients was 2.31 ± 2.87, whereas in ERD patients, it was 0.66 ± 1.11. Despite the fact that the relative expression of cyclin A mRNA in NERD epithelium was fourfold higher than in ERD epithelium, the difference between the two groups was not statistically significant (P = 0.158); both for the low number of cases evaluated, in particular in the ERD group, and for the high variability of the values relative to NERD patients.
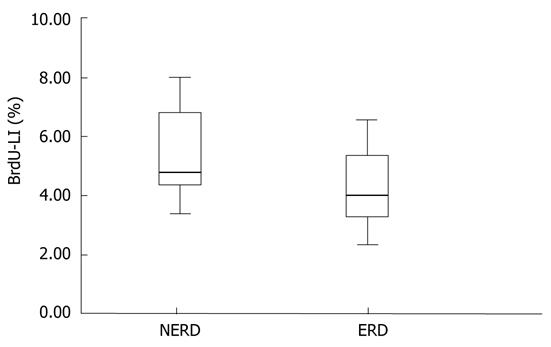
Twelve patients were evaluable for BrdU analysis. BrdU-LI ranged from 2.33% to 8%, with a mean (± SD) value of 4.95% ± 1.67%. The mean BrdU-LI of the NERD patients (n = 7) was 5.42 % ± 1.68%, whereas in ERD patients (n = 5), it was 4.3% ± 1.59% (Figure 5). Once again, NERD epithelium showed a greater number of BrdU-positive cells than ERD epithelium did, but the difference between the two groups was not statistically significant (P = 0.272).
In the present study, we evaluated a series of esophageal biopsies to define the proliferation activity of the epithelium in patients with erosive or non-erosive GERD. In previous investigations, we have demonstrated that, in patients with GERD, cell proliferation evaluated by MIB1 immunostaining was reduced in esophageal mucosa exposed to chronic acid-peptic insult[2,3]. In particular, patients with NERD and ERD showed a decrease in cell proliferation to 50% and 75%, respectively, compared to normal subjects[2].
In contrast to our results, Mastracci et al[4] have found that MIB1 immunostaining of GERD patients is significantly greater than in controls. These different data might reflect different sampling conditions that could influence the proliferating activity of the epithelial cells. In particular, Mastracci and co-workers have evaluated specimens that were taken from 2-4 cm to the Z line, and observed a progressive decrease in the Ki-67 LI by increasing the distance from the Z line.
Feagins et al[10] have shown that multiple acid exposures decrease cell proliferation in non-neoplastic, telomerase-immortalized Barrett’s cell lines. This decrease in cell proliferation is the result of a delay in cell cycle progression that is mediated by p53. In agreement with these results, we have recently demonstrated that, in patients with ERD and NERD, long-term PPI therapy increases esophageal cell proliferation[3]. These data confirm that acid-peptic insults have an antiproliferative effect on esophageal epithelial cells.
In the present study, only patients with at least a 1-year history of GERD were included. Upper endoscopy was performed and biopsies were taken only in apparently normal mucosa at 5 cm above the Z-line. In this way, we studied the behavior of the mucosa exposed to chronic acid insult, but far from erosions, and especially, from reparative changes secondary to the lack of the superficial mucosa, where basal cell hyperplasia has been reported[11], which can be characterized by increased proliferative activity.
Regardless of these considerations, in the present study, we evaluated proliferative activity of the epithelium in patients with erosive and non-erosive GERD. For this purpose, three proliferation markers were assessed: cyclins A and D relative expression, evaluated by real-time RT-PCR, and in vitro BrdU incorporation for immunohistochemical detection of S-phase cells in histological samples.
Cyclins are a family of proteins involved in cell cycle regulation. Cyclin expression rises and falls at various stages of the cell cycle, thus activating specific cyclin dependent kinases (CDKs), which, by phosphorylation of multiple substrates, control a number of critical steps in cell cycle progression[12]. Cyclin D1 is encoded by the CCND1 gene located on chromosome 11q13, and in association with CDK4 or CDK6, regulates the transition from G1 to S phase[13,14]. It is synthesized in response to extracellular mitogenic signals and is maximally expressed in mid-to-late G1-phase[15]. Cyclin A, in association with CDK1 or CDK2, promotes the transition from G2 to M phase[16], and is expressed later in the cell cycle, during DNA replication, achieving its maximal levels during late S-phase[17,18]. Cyclins D1 and A are regarded as specific markers of the G1 and S phases of the cell cycle, respectively. Therefore, in the present study, we evaluated their mRNA expression to assess the proliferative activity of esophageal epithelial cells. In previous investigations, we have in fact demonstrated that the relative expression of cyclin mRNA is directly related to the cell proliferation rate in breast cancer specimens[19]. Moreover, to identify S-phase cells specifically, in the present study, DNA-synthesizing cells were detected in situ after BrdU incorporation by immunohistochemical analysis with anti-BrdU antibodies.
Our results demonstrated that cyclin D, as a marker of G1 phase, was significantly higher in NERD compared to ERD. Also the S-phase markers evaluated in our study (cyclin A and BrdU) were higher in NERD compared to ERD, although in this case, because of the small number of evaluable samples, the difference was not statistically significant. The reduction in the number of samples analyzed for cyclin A compared to those analyzed for cyclin D was due to the fact that the thickness of the epithelium in ERD was significantly reduced, and therefore, in these samples, it was not always possible to isolate a sufficient number of epithelial cells for molecular analysis.
The limitation of this study was the small number of patients, but this is believed to be the first study to evaluate, at the molecular level, esophageal epithelial cells. This method is clean but it creates considerable tissue loss.
In conclusion, the present study confirmed our previous results regarding the reduction of epithelial proliferative activity in ERD compared with NERD patients. Besides, this study supports the previous data on an antiproliferative effect of acid-peptic injury in esophageal cell epithelium, and reinforces the idea that individuals who develop ERD might be genetically characterized by weaker epithelial cell proliferation. On the other hand, patients with more efficient epithelial proliferation capability could have a lower probability of developing macroscopic mucosal lesions when stressed by acid and pepsin. Further studies are required to understand better the mucosal defense mechanisms, and in particular, those controlling the cellular proliferative activity of esophageal mucosa.
Cell replication in basal layers has been suggested as one of the causes of mucosal resistance and structural epithelial defense. To elucidate better the different proliferative activity between non-erosive reflux disease (NERD) and erosive reflux disease (ERD), the authors evaluated a series of molecular and immunohistochemical markers of cell proliferation in squamous epithelial cells of patients with gastroesophageal reflux disease (GERD).
In previous investigations, the authors have demonstrated that, in patients with GERD, cell proliferation evaluated by MIB1 immunostaining is reduced in esophageal mucosa exposed to chronic acid-peptic insult, in contrast with other studies. These different data might reflect different sampling conditions that could influence the proliferating activity of the epithelial cells.
The present study confirmed the previous results with regard to reduction of epithelial cell proliferative activity in ERD compared with NERD patients. It supports previous data on an antiproliferative effect of acid-peptic injury in esophageal cell epithelium, and reinforces the idea that individuals who develop ERD might be genetically characterized by a weaker proliferating epithelial cell capability.
This paper shows that patients with more efficient epithelial proliferative capability could have a lower probability of developing macroscopic mucosal lesions when stressed by acid and pepsin.
The paper by Dr. Calabrese and colleagues discusses activity of cells in esophageal squamous epithelium in patients with NERD and ERD. This is a very well written manuscript that contributes to the knowledge of the molecular mechanisms in patients with GERD.
Peer reviewer: Wojciech Blonski, MD, PhD, University of Pennsylvania, GI Research-Ground Centrex, 3400 Spruce St, Philadelphia, PA 19104, United States
S- Editor Tian L L- Editor Kerr C E- Editor Zhang DN
| 1. | Hershcovici T, Fass R. Nonerosive Reflux Disease (NERD) - An Update. J Neurogastroenterol Motil. 2010;16:8-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 2. | Calabrese C, Trerè D, Fabbri A, Cenacchi G, Vici M, Derenzini M, Di Febo G. Endoscopic appearance of GERD: putative role of cell proliferation. Dig Liver Dis. 2007;39:713-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Calabrese C, Treré D, Liguori G, Gabusi V, Vici M, Cenacchi G, Derenzini M, Di Febo G. Esophageal cell proliferation in gastroesophageal reflux disease: clinical-morphological data before and after pantoprazole. World J Gastroenterol. 2009;15:936-941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Mastracci L, Grillo F, Zentilin P, Spaggiari P, Dulbecco P, Pigozzi S, Savarino V, Fiocca R. Cell proliferation of squamous epithelium in gastro-oesophageal reflux disease: correlations with clinical, endoscopic and morphological data. Aliment Pharmacol Ther. 2007;25:637-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Pacini F, Calabrese C, Cipolletta L, Valva MD, Russo A, Savarino V, Vigneri S. Burden of illness in Italian patients with gastro-oesophageal reflux disease. Curr Med Res Opin. 2005;21:495-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Johnson LF, Demeester TR. Twenty-four-hour pH monitoring of the distal esophagus. A quantitative measure of gastroesophageal reflux. Am J Gastroenterol. 1974;62:325-332. [PubMed] |
| 7. | Weusten BL, Roelofs JM, Akkermans LM, Van Berge-Henegouwen GP, Smout AJ. The symptom-association probability: an improved method for symptom analysis of 24-hour esophageal pH data. Gastroenterology. 1994;107:1741-1745. [PubMed] |
| 8. | Armstrong D, Bennett JR, Blum AL, Dent J, De Dombal FT, Galmiche JP, Lundell L, Margulies M, Richter JE, Spechler SJ. The endoscopic assessment of esophagitis: a progress report on observer agreement. Gastroenterology. 1996;111:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 779] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 9. | Ismail-Beigi F, Horton PF, Pope CE. Histological consequences of gastroesophageal reflux in man. Gastroenterology. 1970;58:163-174. [PubMed] |
| 10. | Feagins LA, Zhang HY, Hormi-Carver K, Quinones MH, Thomas D, Zhang X, Terada LS, Spechler SJ, Ramirez RD, Souza RF. Acid has antiproliferative effects in nonneoplastic Barrett's epithelial cells. Am J Gastroenterol. 2007;102:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Funch-Jensen P, Kock K, Christensen LA, Fallingborg J, Kjaergaard JJ, Andersen SP, Teglbjaerg PS. Microscopic appearance of the esophageal mucosa in a consecutive series of patients submitted to upper endoscopy. Correlation with gastroesophageal reflux symptoms and macroscopic findings. Scand J Gastroenterol. 1986;21:65-69. [PubMed] |
| 12. | Cordon-Cardo C. Mutations of cell cycle regulators. Biological and clinical implications for human neoplasia. Am J Pathol. 1995;147:545-560. [PubMed] |
| 13. | Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1159] [Cited by in RCA: 1245] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 14. | Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689-3695. [PubMed] |
| 15. | Sherr CJ. Cancer cell cycles. Science. 1996;274:1672-1677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3965] [Cited by in RCA: 4006] [Article Influence: 138.1] [Reference Citation Analysis (0)] |
| 16. | Fotedar R, Fotedar A. Cell cycle control of DNA replication. Prog Cell Cycle Res. 1995;1:73-89. [PubMed] |
| 17. | Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 610] [Cited by in RCA: 669] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 18. | Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 540] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 19. | Montanaro L, Vici M, Donati G, Ceccarelli C, Santini D, Treré D, Derenzini M. Controversial relationship between the expression of the RB pathway components and RB protein phosphorylation in human breast cancer. Histol Histopathol. 2007;22:769-775. [PubMed] |