Published online Oct 7, 2011. doi: 10.3748/wjg.v17.i37.4225
Revised: March 24, 2011
Accepted: March 31, 2011
Published online: October 7, 2011
AIM: To identify the novel methylation-silenced gene pentraxin 3 (PTX3) in esophageal squamous cell carcinoma (ESCC).
METHODS: PTX3 mRNA expression was examined in six human ESCC cell lines, one human immortalized normal esophageal epithelial cell line, primary ESCC tumor tissue, and paired adjacent nontumor tissue using reverse transcription polymerase chain reaction (RT-PCR). Semi-quantitative immunohistochemistry was used to examine cellular localisation and protein levels. Methylation specific PCR and bisulphite genomic sequencing were employed to investigate the methylation of the candidate gene.
RESULTS: In the majority of ESCC cell lines, we found that PTX3 expression was down-regulated due to gene promoter hypermethylation, which was further confirmed by bisulphite genomic sequencing. Demethylation treatment with 5-aza-2’-deoxycytidine restored PTX3 mRNA expression in ESCC cell lines. Methylation was more common in tumor tissues (85%) than in adjacent nontumor tissues (25%) (P < 0 .01).
CONCLUSION: PTX3 is down-regulated through promoter hypermethylation in ESCC, and could potentially serve as a biomarker of ESCC.
-
Citation: Wang JX, He YL, Zhu ST, Yang S, Zhang ST. Aberrant methylation of the 3q25 tumor suppressor gene
PTX3 in human esophageal squamous cell carcinoma. World J Gastroenterol 2011; 17(37): 4225-4230 - URL: https://www.wjgnet.com/1007-9327/full/v17/i37/4225.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i37.4225
Esophageal cancer is the sixth most common cause of cancer death worldwide, with over 400 000 new cases diagnosed each year[1]. Esophageal squamous cell carcinoma (ESCC) has a high morbidity and mortality rate in China. However, the molecular mechanisms underlying ESCC development remain poorly understood.
Human carcinogenesis is a multi-stage process in which genetic and epigenetic changes lead to oncogene activation and tumor suppressor gene inactivation[2]. Epigenetic changes, such as promoter DNA methylation, can induce the inactivation of tumor suppressor genes. DNA methylation plays a crucial role in the development of nearly all types of cancer[3]. Recently, a growing list of aberrantly methylated genes has been reported in ESCC, including esophageal cancer related gene 4[4], p16[5], adenomatous polyposis coli[6], transmembrane protein endothelial factor[7], deleted in liver cancer 1[8], ubiquitin carboxy-terminal hydrolase 1[9], testis-specific Y-like protein 5, and human protein phosphatase-1 regulatory subunit-14A[10]. Nevertheless, most of these tumor suppressor genes exhibit a relatively low frequency of methylation in ESCC. Thus, further studies of a greater number of genes involved in the disease pathogenesis and progression are needed to identify putative epigenetic biomarkers for this tumor type.
The pentraxin 3 (PTX3) gene at 3q25 is a member of the pentraxin superfamily. PTX3 expression is induced in response to inflammatory signals, and is produced at the site of inflammation by several cell types, primarily monocytes/macrophages, dendritic cells (DCs), endothelial cells, smooth muscle cells (SMCs), and fibroblasts. PTX3 can combine with a variety of soluble receptor ligands, and plays multiple biological roles, such as immune defense, female reproductive fertility, atherosclerosis, apoptosis, and the regulation of angiogenesis[11-14]. To date, there has been no reported study concerning PTX3 gene promoter methylation.
In the present study, we examined reactivation of epigenetically silenced genes using an oligonucleotide microarray in ESCC cell lines. We also investigated the gene expression profiles of tumor tissue and nontumor tissue in ESCC. The genes markedly up-regulated by 5-aza-2’-deoxycytidine (5-Aza-dC) treatment in an ESCC cell line and markedly decreased in tumor tissue compared with nontumor tissue were considered genes of interest. Bisulphite sequencing and methylation-specific polymerase chain reaction (MSP) analyses were carried out on these genes to confirm the presence of aberrantly methylated CpG dinucleotides. Using the methods mentioned above, we successfully identified PTX3 as a new epigenetically silenced hypermethylated gene in ESCC.
Six human ESCC cell lines were utilized in this study (TE-11, KYSE-30, KYSE-410, KYSE-510, EC-109, and EC-9706) [from American type culture collection (ATCC) and Sciencell]. One human immortalized normal esophageal epithelial cell line (Het-1A) (from ATCC) was used as the “normal” control for ESCC. The ESCC cell lines were cultured in Roswell Park Memorial Institute 1640 supplemented with 10% fetal bovine serum (Hyclone, United States) and antibiotics (100 U/mL penicillin G and 100 μg/mL streptomycin) at 37 °C in a humidified 5% CO2 incubator. Het-1A cells were maintained in bronchial epithelial basal media with growth supplements (Clonetics, United States).
Twenty primary ESCC and paired adjacent nontumor tissues were obtained from the Beijing Friendship Hospital, Beijing, China. Specimens were snap-frozen in liquid nitrogen and subsequently stored at -80 °C. Formalin-fixed, paraffin-embedded samples of 79 primary ESCC cancer tissue specimens and paired adjacent nontumor tissues were also obtained from the Beijing Friendship Hospital. All patients from whom we obtained the study specimens gave informed consent to participate in this study. All case samples were collected from the primary surgical resection in patients with no prior history of ESCC and adjuvant therapy. Pathological diagnosis was performed and confirmed in the Pathology Department. Tumors were histopathologically classified according to TNM (tumor node metastasis) criteria.
ESCC cell lines and Het-1A were treated with 10 μmol/L of the DNA demethylating agent 5-Aza-dC (Sigma-Aldrich, United States) for 4 d.
Total RNA was extracted from cell line pellets and tissues using Trizol (Invitrogen, United States). Reverse transcription was performed using total RNA (1 μg) with Reverse Transcription System (Applied Biosystems, United States). The PTX3 mRNA expression levels were detected by conventional RT-PCR with Taq polymerase (Takara, Japan). RT-PCR was performed for 35 cycles at an annealing temperature of 55 °C. Glyceraldehyde-3-phosohate dehydrogenase (GAPDH) was used as an internal control of RNA integrity. Primer sequences for PTX3 are as follows: (1) PTX3-F: 5’- TCTCTGGTCTGCAGTGTTGG-3’; (2) PTX3-R: 5’- TGAAGAGCTTGTCCCATTCC-3’; (3) GAPDH-F: 5’-CGGAGTCAACGGATTGGTCGTAT -3’; and (4)GAPDH-R: 5’-AGCCTTCTCCATGGTGGTGAAGAC-3’.
Genomic DNA was extracted from cells and tissues by standard phenol–chloroform extraction. Bisulphite modification of DNA was produced with a Zymo DNA Modification Kit (Zymo Research, United States) according to the manufacturer’s protocol.
Bisulphite-treated DNA was amplified with the methylation-specific primer set, PTX3-MF: 5’-CGTTTGCGGTTAGGAGTATTC-3’, and PTX3-MR: 5-CAAAACGTCGTCCGTAACTTA-3’, or the unmethylation-specific primer set, PTX3-UF: 5’-TGTGTTTGTGGTTAGGAGTATTTG-3’ and GPX3-UR: 5’- CAAAACATCATCCATAACTTA-3’, in a total volume of 20 μL using a 0.5 unit of hot-start Taq-polymerase (Takara, Japan) per reaction. The size of the unmethylated amplicon is 105 bp, and the methylated amplicon is 103 bp. Cycle conditions were: initial denaturation and hot start for 10 min at 95 °C, then 35 cycles consisting of 30 s at 94 °C, 30 s at 59 °C (for methylated reactions) or 50 °C (for unmethylated reactions), and 45 s at 72 °C, and a final extension of 10 min at 72 °C.
For the bisulphite genomic sequencing technique, the targeted fragment was amplified from bisulphite-treated DNA, cloned, and sequenced to obtain an accurate map of the distribution of CpG methylation. Forward: 5’TTTTTGAGATATTTATATGTTGTTTTT-3’ and Reverse: 5’AAACACTAATCAACCTAACCTCTAC-3’. PCR was performed for 40 cycles at an annealing temperature of 54 °C. The PCR products were then cloned into the pEasy-T1 vector (Transgene, China), and eight to ten colonies were randomly chosen and sequenced.
Expression of PTX3 was investigated in 79 primary ESCC cancer tissue specimens and in paired adjacent nontumor tissues using immunohistochemistry. The immunohistochemical expression of PTX3 was examined with light microscopy. The percentage of positive tumor cells was determined semi-quantitatively by assessing the entire tumor section, and each sample was assigned to one of the following categories: 0 (0%-4%), 1 (5%-24%), 2 (25%-49%), 3 (50%-74%), or 4 (75%-100%)[15]. The intensity of the immunostain was categorised as 0 (no stain), 1+ (weak stain), 2+ (medium stain), and 3+ (strong stain). Additionally, an immunoreactive score was calculated by multiplying the percentage of positive cells and the stain intensity. In instances of heterogeneous staining intensity within one sample, each component was scored independently and the results were summed. Using this system, the maximum score was 12. For statistical analysis, the criteria were combined as follows: 0, negative; 1 to 4, weakly positive; 5 to 8, moderately positive; and 9 to 12, strongly positive[16].
Statistical analysis was performed using the Student’s t test and the χ2 test; a value of P < 0.05 was considered to be statistically significant.
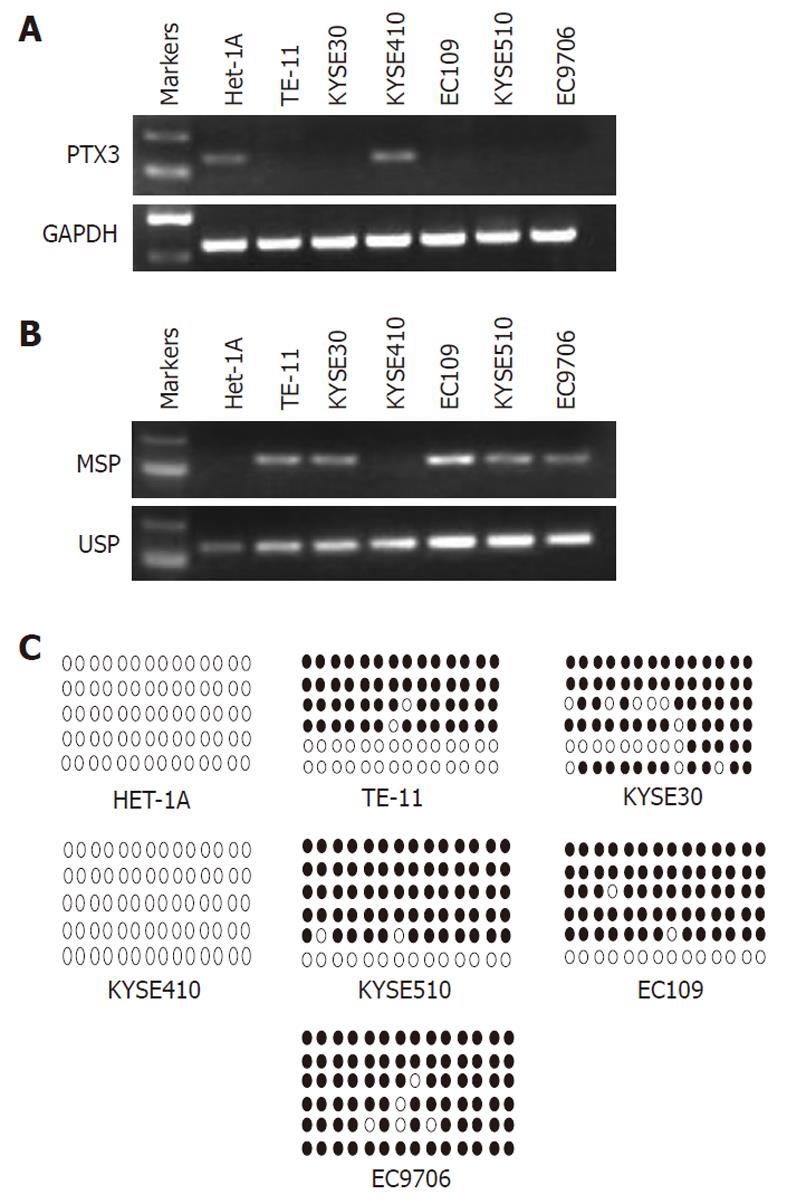
Using RT-PCR with glyceraldehyde 3-phosphate dehydrogenase as a control, we tested the mRNA expression of PTX3 in the immortalized normal esophageal epithelial cell line Het-1A, and in 6 ESCC cell lines (TE-11, KYSE-30, KYSE-410, KYSE-510, EC-109, and EC-9706). The PTX3 transcript was silenced in 5 ESCC cell lines (TE-11, EC109, EC9706, KYSE30, and KYSE510), but not in KYSE-410 and Het-1A (Figure 1A).
As DNA methylation is an important regulator at the transcription level, we hypothesized that the silencing of PTX3 could be caused by methylation. Thus, we performed the MSP assay to measure the methylation level in these ESCC cell lines. The MSP results indicated that partial methylation was observed in the ESCC cell lines TE-11, EC109, EC9706, KYSE30, and KYSE510, whereas the unmethylated promoter was detected in KYSE-410 and in the immortalized normal esophageal epithelial cell line (Het-1A) (Figure 1B). The methylation results were in concordance with the transcription results of these ESCC cell lines.
We performed bisulphite genomic sequencing analysis to confirm the MSP result, and as shown in Figure 1C, the BGS result was consistent with our MSP data: no methylated CpG sites detected in KYSE-410 and Het-1A, but methylated CpG sites in the other ESCC cell lines (TE-11, EC109, EC9706, KYSE30 and KYSE510). The results indicated that transcriptional silencing of PTX3 was associated with methylation in ESCC cells.
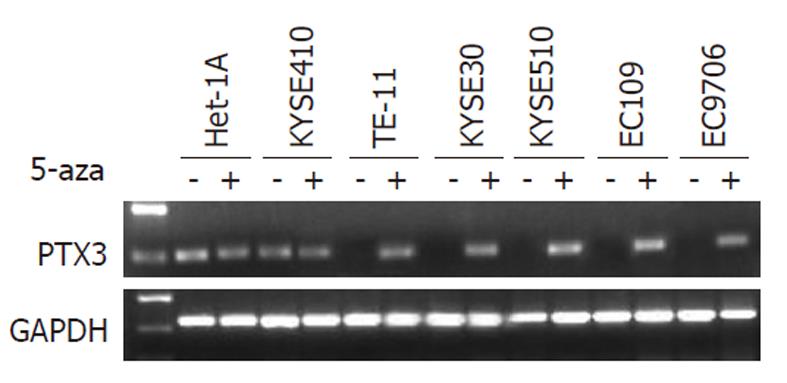
The results of RT-PCR indicated that five ESCC cell lines did not express PTX3 mRNA (Figure 1A). As mentioned above, there was a correlation between PTX3 silencing and DNA methylation in ESCC cell lines. To determine whether PTX3 expression could be reactivated by pharmacological demethylation of genomic DNA, all cell lines above were treated with the demethylating agent 5-Aza-dC. After treatment with 5-Aza-dC at 10 μmol/L for 4 d, the five silenced ESCC cell lines resulted in an obvious increase in PTX3 expression (Figure 2), further supporting the role of methylation as a primary mechanism of PTX3 silencing.
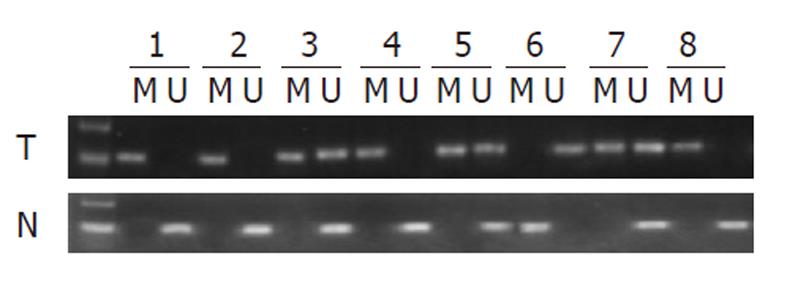
To assess whether the PTX3 promoter hypermethylation observed in cell lines was relevant to ESCC, we further examined PTX3 methylation in 20 primary ESCC tumors with paired adjacent nontumor tissues using MSP. We found that 80% of ESCC tumor samples (16 of 20) exhibited statistically different methylation within the PTX3 promoter region, whereas the paired nontumor tissues exhibited only 20% (5 of 20) (P < 0.001).
The MSP results of ESCC primary tumors (T) and paired adjacent nontumor tissues (N) are shown in Figure 3.
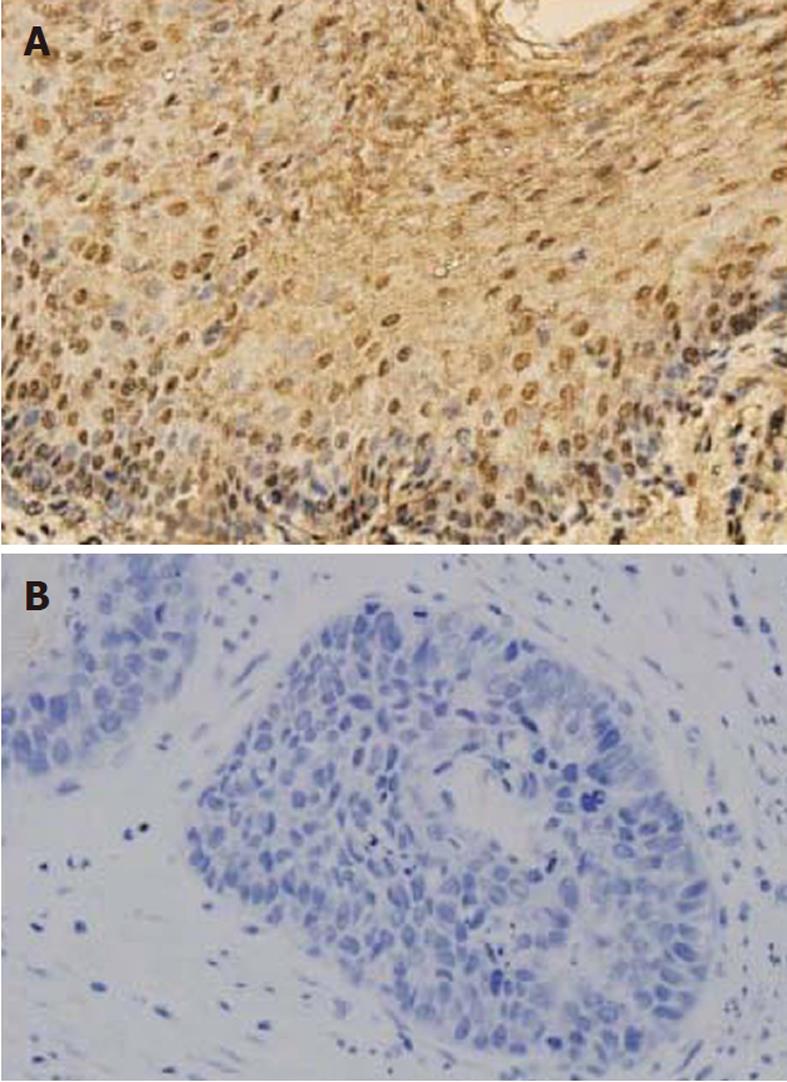
We then analyzed 79 primary ESCC specimens and their corresponding adjacent nontumor tissues using immunohistochemical staining. PTX3 protein was detected in 24 of 79 (30.38%) ESCC specimens. In the non-malignant tissues, 67 of 79 (84.81%) samples showed positive detection of PTX3 protein.
In adjacent nontumor tissues, intense immunostaining for PTX3 was observed consistent with cytoplasmic distribution (Figure 4), whereas absent or weak immunostaining was detected in tumor tissues. Immunohistochemical results revealed that the expression of PTX3 protein in ESCC tumor tissues was significantly lower compared to adjacent nontumor tissues (P < 0.01).
The clinicopathological features of these patients and the results of PTX3 expression are summarized in Table 1. Statistical analysis indicated that PTX3 protein expression exhibited no correlation with the patients’ age, gender, smoking habit, depth of invasion, or lymph node metastasis (P > 0.05). However, we found a higher frequency of promoter hypermethylation of PTX3 in the early stages of cancer (Iand II) compared to advanced stages, suggesting that PTX3 hypermethylation occurs during a relatively early stage of the multi-step esophageal carcinogenesis.
| Clinicopathological features | PTX3 expression | ||
| Positive | Negative | P value | |
| Cases | 24 (30) | 55 (70) | |
| Gender | |||
| Male | 16 (30) | 38 (70) | 0.831 |
| Female | 8 (32) | 17 (68) | |
| Age (mean, yr) | 59.6 | 61.2 | 0.510 |
| Smoker | 12 (32) | 26 (68) | 0.823 |
| Non-smoker | 12 (29) | 29 (71) | |
| Depth of invasion | 0.315 | ||
| T1, T2 | 16 (35) | 30 (65) | |
| T3, T4 | 8 (24) | 25 (76) | |
| Lymph node metastasis | 0.311 | ||
| Positive | 4 (20) | 16 (80) | |
| Negative | 20 (34) | 39 (66) | |
| TNM classification | 0.012 | ||
| I | 9 (23) | 20 (77) | |
| IIa | 10 (35) | 19 (65) | |
| IIb | 2 (29) | 5 (71) | |
| III | 1 (11) | 8 (89) | |
| IV | 2 (67) | 3 (23) | |
Tumorigenesis is a multistep process caused by the accumulation and interplay of genetic and epigenetic alterations. DNA methylation is a key regulator of gene transcription and genomic stability. Alteration of DNA methylation is one of the most consistent epigenetic changes that silence tumor suppressor genes in human cancers[17]. Additionally, aberrant methylation results in increased gene mutagenicity, due to the deamination of 5-methylcytosine to thymine[17].
We have previously analyzed expression microarray data prior to and post treatment using a demethylating agent in the EC9706 cell line, and we speculated that the PTX3 gene is potentially down-regulated by promoter hypermethylation. PTX3 is involved in the regulation of innate resistance to pathogens, the inflammatory reaction, and possibly the clearance of self-components and female fertility. Transfection of PTX3 into breast cancer cells lacking expression led to a reduction in endothelial cell invasion and capillary tube formation, as well as prevention of tumor formation in athymic nude mice[18]. In this study, we examined the methylation status of the PTX3 promoter region in ESCC. As far as we understand, this is the first study to report on PTX3 gene promoter methylation in ESCC.
Down-regulation of PTX3 mRNA and protein was detected by RT-PCR and immunostaining. We found that PTX3 was silenced in most ESCC cell lines (5 of 6 ESCC cell lines that we tested) and also silenced in tumor tissues. Reduced expression of PTX3 suggests that PTX3 plays a tumor suppressive role in ESCC.
PTX3 promoter hypermethylation was confirmed by methylation specific PCR and bisulphate genomic sequencing methylation in 83.3% of ESCC cell lines (5/6) and 80% of primary esophageal squamous cell carcinoma tissues (16/20), suggesting that promoter hypermethylation is a major, if not the only, mechanism for PTX3 down-regulation in ESCC.
Treatment of ESCC cell lines in vitro with 5-Aza-dC, a nucleoside analogue inhibitor of DNA methyltransferase, reversed PTX3 CpG island hypermethylation and restored PTX3 expression; this confirmed that PTX3 hypermethylation serves as the principal mechanism for PTX3 downregulation in ESCC. PTX3 hypermethylation is involved in the development and progression of esophageal cancer.
In our study, PTX3 protein expression was not associated with patients’ age, gender, smoking habit, depth of invasion, and lymph node metastasis. We found a high frequency of promoter hypermethylation of PTX3 in the early tumor stages (I and II) of ESCC, indicating that aberrant methylation is a relatively early event in esophageal carcinogenesis.
Methylation-mediated inactivation is reversible, and up-regulation of PTX3 by 5-aza-dC may reverse the malignant phenotype of tumor cells. Therefore, PTX3 could serve as a novel target for gene therapy in ESCC treatment. In the future, further studies to elucidate the function of PTX3 in ESCC are warranted.
In summary, our study provides the first documentation that PTX3 is a novel tumor suppressor gene epigenetically silenced in most ESCC tumors. Our results suggest that methylation of the PTX3 promoter region occurs at an early stage of ESCC pathogenesis and may provide a suitable biomarker for ESCC diagnosis.
Esophageal cancer is the sixth most common cause of cancer death worldwide, with > 400 000 new cases diagnosed each year. The molecular mechanisms underlying esophageal squamous cell carcinoma (ESCC) development remain poorly understood. DNA methylation plays a crucial role in the development of ESCC.
Oligonucleotide Microarray Analysis can be used to identify novel genes that are aberrantly methylated in ESCC. Genes that were significantly upregulated after 5-aza-2’-deoxycytidine (5-Aza-dC) treatment in ESCC cells and significantly downregulated in tumor tissue compared with paired nontumor tissue were selected as hypermethylated candidate genes. Subsequently, bisulphite sequencing and methylation-specific PCR were performed to confirm the presence of aberrantly methylated CpG dinucleotides. In this study, the authors successfully identified pentraxin 3 (PTX3) as a new methylation-silenced gene in ESCC.
The study provides the first documentation that PTX3 is a novel candidate tumor suppressor gene epigenetically silenced in most ESCC tumors. The results suggest that methylation of the PTX3 promoter region occurs at an early stage of ESCC pathogenesis and may be used as a biomarker for ESCC diagnosis.
Up-regulating PTX3 by 5-aza-dC may reverse the malignant phenotype of tumor cells. Therefore, PTX3 could be used as a novel target for gene therapy in ESCC treatment. PTX3 functions extracellularly as a secreted protein. PTX3 plays a tumor suppressive role in ESCC; thus, PTX3 protein could potentially be used directly as an anticancer drug therapy.
PTX3 gene at 3q25 is a member of the pentraxin super-family. PTX3 expression is induced in response to inflammatory signals. PTX3 is able to combine with a variety of soluble receptor ligands and play multiple biological roles, such as immune defense, female reproductive fertility, atherosclerosis, apoptosis, and regulation of angiogenesis.
This study reports the newly recognized tumor suppressor features of the gene PTX3, a gene that encodes a protein referred to as pentraxin 3 that serves as a protector against pathogens in tissues and aids in the control of autoimmunity. As an acute phase reactant, its blood level increases significantly during sepsis and severe inflammation, correlating with the severity of the disease and making the protein a useful biomarker for many inflammatory diseases. The authors of this study suggest that PTX3 could also be utilized as a tumor marker in ESCC.
Peer reviewer: Mehmet Fatih CAN, Assistant Professor, De-partment of General Surgery, Gulhane School of Medicine, Gulhane Askeri Tıp Akademisi, Genel Cerrahi AD, Etlik, Ankara, 06018, Turkey
S- Editor Sun H L- Editor O’Neill M E- Editor Xiong L
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13286] [Cited by in RCA: 13558] [Article Influence: 677.9] [Reference Citation Analysis (1)] |
| 2. | Ponder BA. Cancer genetics. Nature. 2001;411:336-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 349] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 3. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer. 2009;125:1505-1513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 5. | Salam I, Hussain S, Mir MM, Dar NA, Abdullah S, Siddiqi MA, Lone RA, Zargar SA, Sharma S, Hedau S. Aberrant promoter methylation and reduced expression of p16 gene in esophageal squamous cell carcinoma from Kashmir valley: a high-risk area. Mol Cell Biochem. 2009;332:51-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 6. | Zare M, Jazii FR, Alivand MR, Nasseri NK, Malekzadeh R, Yazdanbod M. Qualitative analysis of Adenomatous Polyposis Coli promoter: hypermethylation, engagement and effects on survival of patients with esophageal cancer in a high risk region of the world, a potential molecular marker. BMC Cancer. 2009;9:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Zhao BJ, Tan SN, Cui Y, Sun DG, Ma X. Aberrant promoter methylation of the TPEF gene in esophageal squamous cell carcinoma. Dis Esophagus. 2008;21:582-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 105] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Yu J, Tao Q, Cheung KF, Jin H, Poon FF, Wang X, Li H, Cheng YY, Röcken C, Ebert MP. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Oka D, Yamashita S, Tomioka T, Nakanishi Y, Kato H, Kaminishi M, Ushijima T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412-3426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Okutani D. [The role of long pentraxin 3, a new inflammatory mediator in inflammatory responses]. Nihon Rinsho Meneki Gakkai Kaishi. 2006;29:107-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Soares AC, Souza DG, Pinho V, Vieira AT, Nicoli JR, Cunha FQ, Mantovani A, Reis LF, Dias AA, Teixeira MM. Dual function of the long pentraxin PTX3 in resistance against pulmonary infection with Klebsiella pneumoniae in transgenic mice. Microbes Infect. 2006;8:1321-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 13. | Muller B, Peri G, Doni A, Torri V, Landmann R, Bottazzi B, Mantovani A. Circulating levels of the long pentraxin PTX3 correlate with severity of infection in critically ill patients. Crit Care Med. 2001;29:1404-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 255] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 14. | Peri G, Introna M, Corradi D, Iacuitti G, Signorini S, Avanzini F, Pizzetti F, Maggioni AP, Moccetti T, Metra M. PTX3, A prototypical long pentraxin, is an early indicator of acute myocardial infarction in humans. Circulation. 2000;102:636-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 314] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 15. | Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 980] [Cited by in RCA: 945] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 16. | Milde-Langosch K, Bamberger AM, Rieck G, Kelp B, Löning T. Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype. Breast Cancer Res Treat. 2001;67:61-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3397] [Cited by in RCA: 3721] [Article Influence: 161.8] [Reference Citation Analysis (0)] |
| 18. | Margheri F, Serratì S, Lapucci A, Anastasia C, Giusti B, Pucci M, Torre E, Bianchini F, Calorini L, Albini A. Systemic sclerosis-endothelial cell antiangiogenic pentraxin 3 and matrix metalloprotease 12 control human breast cancer tumor vascularization and development in mice. Neoplasia. 2009;11:1106-1115. [PubMed] |