Published online Jan 21, 2011. doi: 10.3748/wjg.v17.i3.300
- This article has been corrected.
- See: World J Gastroenterol. Jun 21, 2025; 31(23): 108472
Revised: August 3, 2010
Accepted: August 10, 2010
Published online: January 21, 2011
AIM: To develop a hepatocellular carcinoma (HCC) xenograft model for studying hepatitis C virus (HCV) replication in a mice, and antiviral treatment.
METHODS: We developed a stable S3-green fluorescence protein (GFP) cell line that replicated the GFP-tagged HCV sub-genomic RNA derived from a highly efficient JFH1 virus. S3-GFP replicon cell line was injected subcutaneously into γ-irradiated SCID mice. We showed that the S3-GFP replicon cell line formed human HCC xenografts in SCID mice. Cells were isolated from subcutaneous tumors and then serially passaged multiple times in SCID mice by culturing in growth medium supplemented with G-418. The mouse-adapted S3-GFP replicon cells were implanted subcutaneously and also into the liver of SCID mice via intrasplenic infusion to study the replication of HCV in the HCC xenografts. The tumor model was validated for antiviral testing after intraperitoneal injection of interferon-α (IFN-α).
RESULTS: A highly tumorigenic S3-GFP replicon cell line was developed that formed subcutaneous tumors within 2 wk and diffuse liver metastasis within 4 wk in SCID mice. Replication of HCV in the subcutaneous and liver tumors was confirmed by cell colony assay, detection of the viral RNA by ribonuclease protection assay and real-time quantitative reverse transcription polymerase chain reaction. High-level replication of HCV sub-genomic RNA in the tumor could be visualized by GFP expression using fluorescence microscopy. IFN-α cleared HCV RNA replication in the subcutaneous tumors within 2 wk and 4 wk in the liver tumor model.
CONCLUSION: A non-infectious mouse model allows us to study replication of HCV in subcutaneous and metastatic liver tumors. Clearance of HCV by IFN-α supports use of this model to test other anti-HCV drugs.
- Citation: Hazari S, Hefler HJ, Chandra PK, Poat B, Gunduz F, Ooms T, Wu T, Balart LA, Dash S. Hepatocellular carcinoma xenograft supports HCV replication: A mouse model for evaluating antivirals. World J Gastroenterol 2011; 17(3): 300-312
- URL: https://www.wjgnet.com/1007-9327/full/v17/i3/300.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i3.300
Hepatitis C virus (HCV) is the most common blood-borne infection that affects the liver. The majority of individuals infected with HCV end up with a chronic disease in which the virus replicates in the liver for a long period of time. There are approximately 170 million people currently infected with HCV worldwide[1,2]. The incidence of new HCV infections each year is increasing in developing nations due to blood transfusion from unscreened donors, which makes HCV a significant worldwide public health problem. The standard treatment option for chronic HCV infection is combination of pegylated interferon-α (IFN-α) with ribavirin[3]. However, the majority of chronic HCV patients do not clear the virus infection with this treatment, and these individuals remain at high risk for developing cirrhosis and liver cancer. There is no effective treatment available for liver cirrhosis and cancer, so the development of alternative therapeutic strategies using a small animal model system to cure chronic HCV infection is crucial.
Development of novel antiviral therapies that target the multiple steps in the HCV life cycle, including viral genome replication, assembly, and infection is now possible due to the availability of the infectious, full-length HCV cell culture systems. A number of antiviral strategies have been developed during the past 2 years, including small molecules, anti-sense oligonucleotides, siRNA, ribozymes, and recombinant antibodies that have been designed to inhibit HCV replication in cell culture models[4,5]. Progress in the targeted delivery of these antiviral molecules to inhibit viral replication in the liver is hampered due to the lack of a small animal model for HCV infection. The development of an animal model for HCV infection has been difficult due to the fact that the virus has limited host tropism and can infect only humans and chimpanzees[6,7]. The chimpanzee is the only natural animal model for studying HCV infection[8]. The chimpanzee model has been used in the past to study many aspects of HCV pathobiology and infectivity using cloned viral genomes, and it remains an important animal model. However, it is a difficult and expensive animal model that cannot be used to optimize experiments that deal with the liver-targeted delivery of intracellular treatment approaches such as siRNA or antibodies. Therefore, a small animal model for HCV infection is required to test different experimental therapies developed using HCV cell culture systems. Rodents are the preferred animal models for testing gene delivery experiments because of their size, low cost and short gestation period. Previously, researchers have developed mouse models for HCV infection using transgenic technology or direct transfection of HCV RNA into the liver[9]. Transgenic mice are not models for HCV infection or antiviral testing because of stable integration of HCV cDNA into the mouse chromosome. It will be difficult to differentiate the viral RNA replication from the HCV RNA produced due to cellular transcription in the transgenic animal models. There has been limited success using this transgenic approach because mouse hepatocytes do not support HCV RNA replication or infection[9,10].
To overcome this limitation, four different mouse models have been developed to study experimental HCV infection in vivo: the immunotolerized rat model[11]; the Trimera mouse model[12]; the uPA/SCID mouse model[13-15]; and the Fah-/- Rag-/- IL-2-/- mouse model[16]. The rat model for HCV infection has been developed using immunotolerized rat embryos that can allow transplantation of human hepatoma cell lines that can be infected with HCV. These models have been used to study the complete life cycle of HCV infection, and antiviral testing. In the latter three models, researchers have used highly specialized mouse strains that support transplantation of human hepatocytes into the mouse liver. These models appear to be highly relevant animal models for HCV infection. A number of investigators have been using these models to infect chimeric mice with HCV or hepatitis B virus[13-16] However, these models are relatively complicated to use because they require specialized surgical skills, special strains of mice, use of human hepatocytes, as well as HCV-infected serum samples. Recently, a xenograft mouse tumor model for HCV infection has been described that utilizes the mouse-adapted replicon cell line that contains the luciferase reporter[17,18]. The replication of HCV RNA has been measured in the mouse liver and subcutaneous tumor xenograft using a whole-body real-time imaging system. This model has been used to evaluate the antiviral properties of IFN-α and protease inhibitors. There is a lot of interest in this mouse model because it is less expensive, non-infectious, and can be easily adapted to any research laboratory.
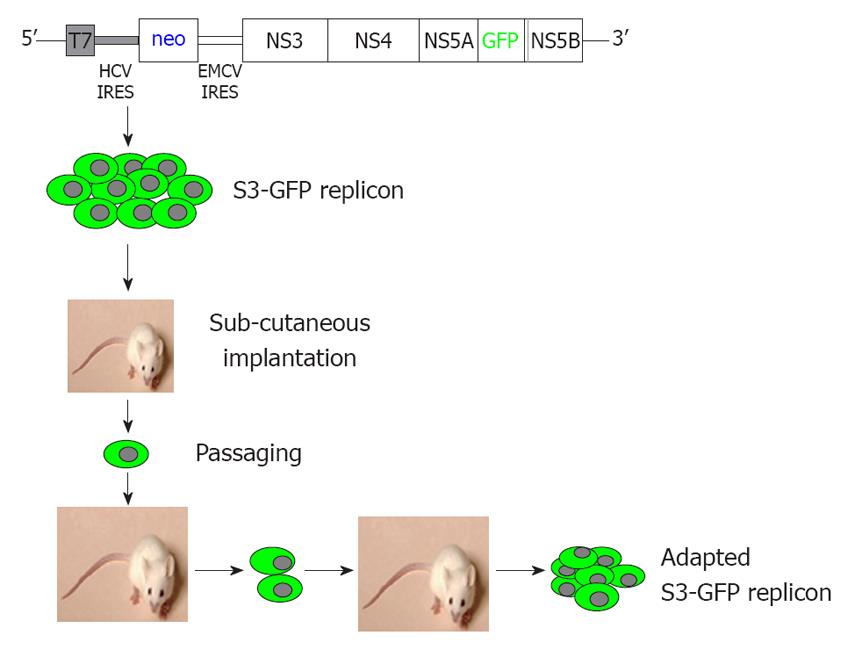
We describe here a mouse model for studying the replication of the JFH1 sub-genomic clone of HCV in subcutaneous and liver tumors using a green fluorescence protein (GFP)-labeled sub-genomic replicon in the Huh-7 cell line. The development of a mouse-adapted GFP-labeled replicon cell line, combined with the highly efficient JFH1 virus facilitates high-level replication of the HCV in subcutaneous and liver tumors of laboratory mice with a partially suppressed immune system. We have developed a mouse-adapted HCC cell clone that replicates sub-genomic HCV RNA and forms HCC xenografts in SCID mice. We show that replication of HCV in the subcutaneous and liver tumors can be assayed using several biochemical methods by looking at GFP expression and evaluating the antiviral effects on HCV by cell colony assay, ribonuclease protection assay, and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR). We showed that IFN-α successfully inhibited replication of HCV RNA in the HCC xenograft, which indicates that the model can be used to test the efficacy of different antiviral strategies that target the HCV replication cycle.
Female SCID/bg and NOD/SCID γ mice 6-8 wk old were obtained from Charles River Laboratories (Wilmington, MA, USA) and Jackson laboratory (Bar Harbor, ME, USA), respectively. The mice were maintained in sterile conditions in a pathogen-free environment at the Department of Comparative Medicine, Tulane University Health Sciences Center. All animal experiments were carried out after receiving approval from the Institutional Animal Care and Use Committee (IACUC), Tulane University Health Sciences Center. All the SCID mice were γ-irradiated at 3 Gy (approximately 3.2 min) 1 d prior to cell transplantation. SCID/bg mice were used for subcutaneous tumor xenografts and NOD/SCID mice were used for liver tumor development. Throughout every experiment, we carefully checked the mice for their well-being, body condition and movement. All of the mice were weighed every alternate day to check for weight loss. A drop in their body weight was considered as an indication of sickness. After surgery, the mice were kept in separate sterile cages and observed carefully until they fully recovered.
We developed a stable Huh-7 cell line (S3-GFP) that replicated the HCV-GFP sub-genomic RNA of the JFH1 clone. The replicon cell line expressed a high level of GFP that could be visualized directly under a fluorescence microscope. We previously have shown that IFN-α inhibits replication of HCV in the S3-GFP cell line in a dose-dependent manner[19]. One million GFP-replicon cells were implanted subcutaneously into the right and left flank of SCID/bg mice. Mice were followed for the development of tumors. When the tumors reached 10 mm in size, they were harvested in a sterile Petri dish. Cells were dissociated by collagen digestion and separated by low-speed (500 rev/min) centrifugation. The cell pellet was resuspended in RBC lysis buffer (eBioscience, San Diego, CA, USA) for 15 min; cells were centrifuged and the cell pellet was resuspended in DMEM with 10% FBS supplemented with G-418 (1 mg/mL). The cells were cultured and expression of GFP in the Huh-7 cells was examined over time under a fluorescence microscope. When most of the Huh-7 cells in the culture showed GFP expression, they were harvested and injected into SCID mice for tumor development. The in vivo passaging experiments were repeated several times until > 50% of the cells in the subcutaneous tumor were GFP-positive.
Intrasplenic infusion of replicon cells was performed using a previously described procedure[20]. NOD/SCID γ mice were anesthetized with isoflurane under a laminar flow cabin. The surgical area was shaved and swabbed with betadine scrub. A small incision was made in the left flank in order to expose the spleen and carry out the cell injection. The spleen was accessed with a small forceps, and 106 replicon cells were injected into the inferior splenic pole; a monofilament suture was placed across the spleen at the site of injection to reduce spillage of cells into the abdominal cavity. The peritoneal wall and skin were separately closed using a monofilament suture and staples, respectively. After 3, 4, 5 and 6 wk, the animals were euthanized by CO2 inhalation and their livers were removed. Part of the liver was fixed in 10% buffered saline for 72 h, processed, and embedded in paraffin. Tissue blocks were made for histological analysis after hematoxylin and eosin staining. The remaining part of the liver tissue was frozen in OCT compound for GFP expression analysis.
IFN-α 2b (Intron A; Schering-Plough, NJ, USA) was diluted in PBS at a concentration of 150 IU/μL and stored at -70°C. Both the subcutaneous and liver tumor models were validated by intraperitoneal injection of 100 μL IFN-α solution (total 15 000 IU/mouse) three times weekly. A group of five mice was used to test the IFN antiviral effect in the subcutaneous and liver tumor models.
The growth of HCC xenografts in the SCID mice was examined by hematoxylin and eosin staining of fixed and paraffin-embedded mouse tumor and liver specimens. Five-micrometer sections were cut from each tissue block, mounted on a glass slide, and dried over night at room temperature. All of the sections were deparaffinized in xylene, rehydrated by dipping in a graded alcohol series, and washed in PBS. To demonstrate the implantation of replicon cells in the liver, the tissue sections were stained with an antibody against human serum albumin (Dako, Carpinteria, CA, USA). The immunoreactivity of the albumin antibody was detected using the ABC detection kit using a standard laboratory protocol. To demonstrate expression of GFP in the subcutaneous and liver tumors, frozen sections were prepared. The sections were washed in PBS and stained with Hoechst dye (H33342, Calbiochem, Germany). Expression of GFP in the HCC xenograft was observed using a fluorescence microscope (Olympus) using a standard procedure[19].
To study the replication of HCV sub-genomic RNA in the HCC xenografts, the liver was digested with collagenase and viable S3-GFP replicon cells were obtained by low-speed centrifugation. The cell pellet was suspended in 5 mL RBC lysis buffer (eBiosciences) for 15 min. The viable tumor cells (S3-GFP) were cultured in DMEM supplemented with G-418 (1 mg/mL). Huh-7 cells that supported HCV RNA replication after IFN treatment were selected. The number of cell colonies formed in each cell culture dish was counted after Giemsa staining (Sigma, St. Louis, MO, USA).
Total RNA was isolated from the subcutaneous tumor by the GITC method and subjected to RPA for the detection of genomic positive-strand HCV RNA, using an anti-sense RNA probe that targeted the 5’ untranslated region (UTR). For the RPA, 25 μg total RNA was mixed with a negative-strand RNA probe that targeted the 5’-UTR of HCV (106 cpm) in a 10-μL hybridization solution, denatured for 3 min at 95°C, and hybridized overnight at 50°C. RNase digestion was performed in 200 μL RNase digestion buffer (10 mmol/L Tris, pH 7.5, 5 mmol/L EDTA and 0.3 mol/L NaCl) that contained RNaseA/T1 cocktail at 1:100 dilutions (Ambion, Austin, TX, USA) for about 1 h at 37°C. It was then treated with 2.5 μL 25% SDS and 10 μL proteinase K (20 mg/mL) for 15 min. Samples were extracted with phenol:chloroform and precipitated with absolute ethanol. The pellet was suspended in 16 μL gel loading buffer, heat denatured, and separated on a 6% TBE-urea gel (Invitrogen, Carlsbad, CA, USA). The gel was dried and exposed to X-ray film (Kodak Biomax-XAR, Rochester, NY, USA). To detect JFH1-HCV mRNA in the transfected cells, we prepared a plasmid construct called pCR-II-2a (Invitrogen), which contained the sequence of 79-297 nt of the 5’-UTR sequence of the JFH1 clone (pCR-II NT-218). This plasmid was linearized with the XbaI restriction enzyme. T7 RNA polymerase was used to prepare a negative-strand RNA probe for detection of positive-strand HCV RNA. The same amounts of the RNA extracts were subjected to RPA for GAPDH mRNA. We used a linearized pTRI-GAPDH-human antisense control template to prepare a probe to detect GAPDH mRNA using Sp6 RNA polymerase (Ambion). The appearance of a 218-nt fragment in the RPA indicated the presence of HCV positive-strand RNA.
To detect HCV RNA in the liver tumors, total RNA was isolated from the liver tissues by the GITC method[21]. cDNA synthesis for HCV positive-strand RNA was carried out using 1 μg total cellular RNA, an outer antisense primer and avian myoblastosis virus (AMV) reverse transcriptase (Promega, Madison, WI, USA). Amplification of the cDNA was performed in 50 μL reaction mixture that contained 250 ng outer sense primer and Taq DNA polymerase. The first PCR product was amplified by another round of PCR using outer and inner sets of primers, as described previously[21]. The HCV PCR products were further subjected to Southern blot analysis by using a positive-sense HCV-specific 32P-labeled oligoprobe. One microgram of total cellular RNA was used to amplify albumin mRNA levels by RT-PCR. The sequence of primer and probe used in these experiments has been described previously[21].
Real time RT-qPCR was performed to quantify HCV RNA levels in the infected cell culture using a published protocol[22]. The 243-bp HCV DNA was amplified from the RNA extract by RT-PCR using the outer sense primer 5'-GCAGAAAGCGCCTAGCCATGGCGT-3' (67-90) and outer antisense primer 5'-CTCGCAAGCGCCCTATCAGGCAGT-3' (287-310). cDNA synthesis was performed from positive-strand HCV RNA using an outer antisense primer that targeted the highly conserved 5’-UTR region of HCV in a 20-μL volume. Two micrograms of total cellular RNA were mixed with 1 μL outer antisense primer (200 ng/μL), denaturized at 65°C for 10 min, and annealed at room temperature. AMV reverse transcriptase (10 U) (Promega) was added and incubated at 42°C for 60 min in the presence of 50 mmol/L Tris, pH 8.3, 50 mmol/L EDTA, 500 nmol/L dNTP, 250 nmol/L spermidine, and 40 U RNasin (Promega). cDNA was stored at -20°C until use. SYBR Green real-time PCR amplification was performed in a 20-μL volume that contained 10 μL SYBR Green ER qPCR SuperMix, 1 μL (250 ng/μL) of sense and antisense primer with 4 μL cDNA and 4 μL distilled water. All samples were run in triplicate. The amplification was carried out using the standard program recommended by Bio-Rad Laboratories that included: a first cycle at 50°C for 2 min, 95°C for 8 min, and an additional 50 cycles in which each cycle consisted of a denaturation step at 95°C for 10 s, and annealing and extension steps at 60°C for 30 s. At the end of the amplification cycles, melting temperature analysis was performed by a slow increase in temperature (0.1°C/s) up to 95°C. Amplification, data acquisition, and analysis were performed on CFX96 Real Time instrument (Bio-Rad Laboratories) using CFX manager software (Bio-Rad Laboratories).
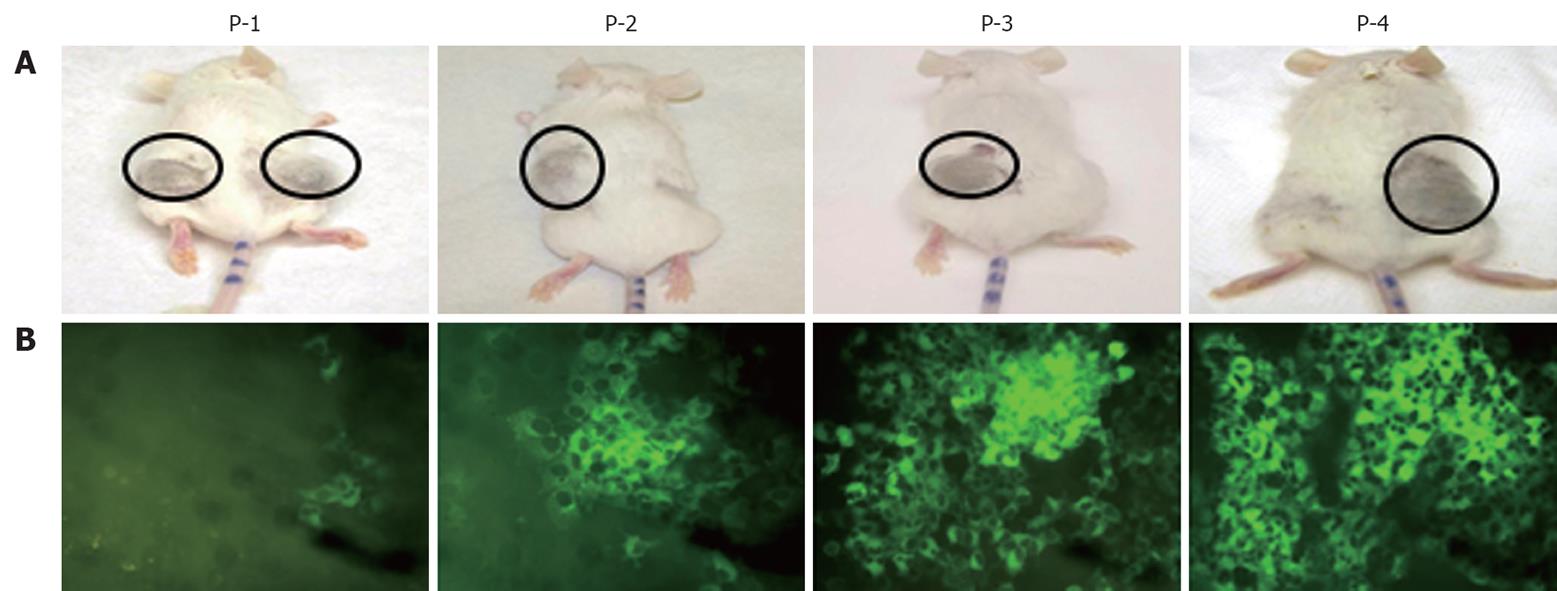
We prepared a chimeric clone of the sub-genomic HCV of JFH1 2a virus by fusing it with GFP in a frame with one of the non-structural proteins of HCV (NS5A). Huh-7 replicon cells that replicated the HCV-GFP sub-genomic RNA emitted a green fluorescent signal when exposed to a specific wavelength of light. This allowed direct visualization of HCV RNA replication. These replicon cells were cultured for > 1 year and were found to have stably maintained GFP expression with > 60%-80% of cells positive by flow analysis. We have shown that IFN-α inhibits replication of HCV RNA and inhibits GFP expression in a dose-dependent manner in the replicon cells[19]. With this GFP-based replicon cell line, we sought to establish an HCC xenograft in SCID mice using the mouse-adapted replicon cells as described previously[18]. The experimental steps involved in generating the in vivo adapted GFP replicon cells are summarized in Figure 1. S3-GFP replicon cells were injected subcutaneously into the right and left flank of SCID mice after γ irradiation. Tumors were harvested when they were 10 mm in size and cells were isolated and cultured in selection medium. After the first passage, we found that expression of GFP in the replicon cells was very faint and only a few cells (< 10%) showed bright GFP positivity. The sub-genomic HCV RNA has a neomycin gene selection marker that allows the selection of tumor cells that support HCV replication. Therefore, replicon cells were cultured with medium that contained G-418, to select for increased intracellular GFP expression. After 1 wk, a high level of GFP expression was seen in most of the cells in culture. This experiment suggested that in vivo tolerance occurred at the cellular level as well as at the level of intracellular virus replication in the replicon cells. We were able to select a homogeneous population of Huh-7 cells with stable GFP expression after the first passage in SCID mice. To obtain a GFP replicon cell line with increased growth and replication capacity, in vivo passaging experiments were repeated three additional times. Figure 2 suggests that in vivo passaging of replicon cells allowed for the selection of replicon cells with increased replicative ability. The expression of GFP in the frozen tumor sections was examined under a fluorescence microscope after nuclear staining. The levels of viral RNA replication in the subcutaneously formed HCC xenografts were very high and GFP expression was observed in most of the replicon cells (Figure 2). We generated a highly adapted GFP replicon cell line (S-3GFP/in vivo) that formed a HCC xenograft in SCID/bg mice within 2 wk.
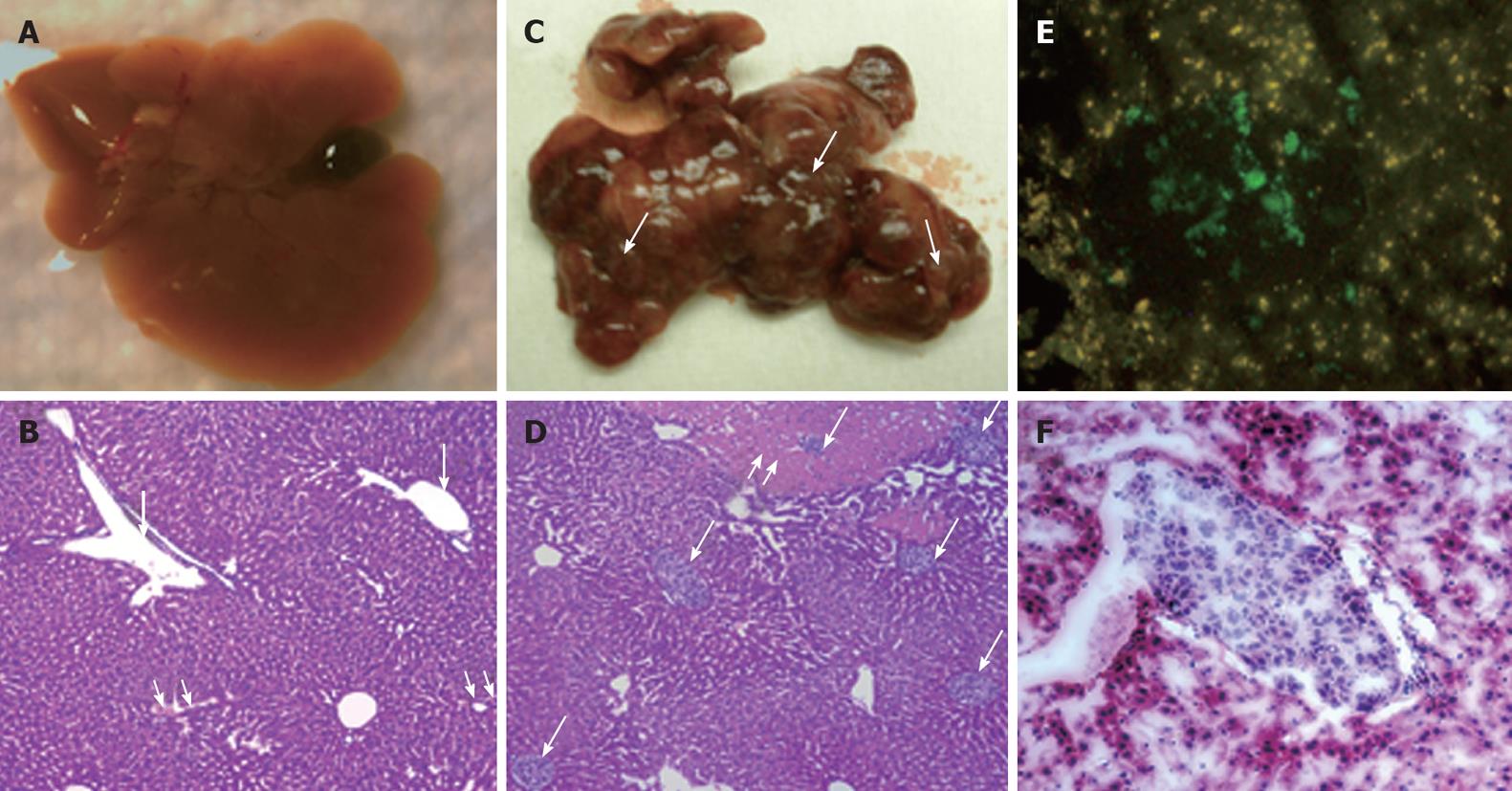
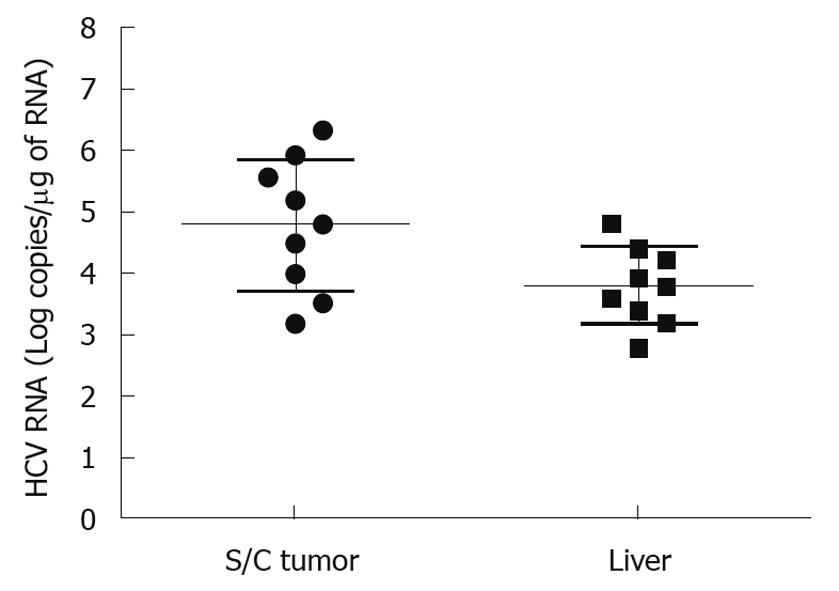
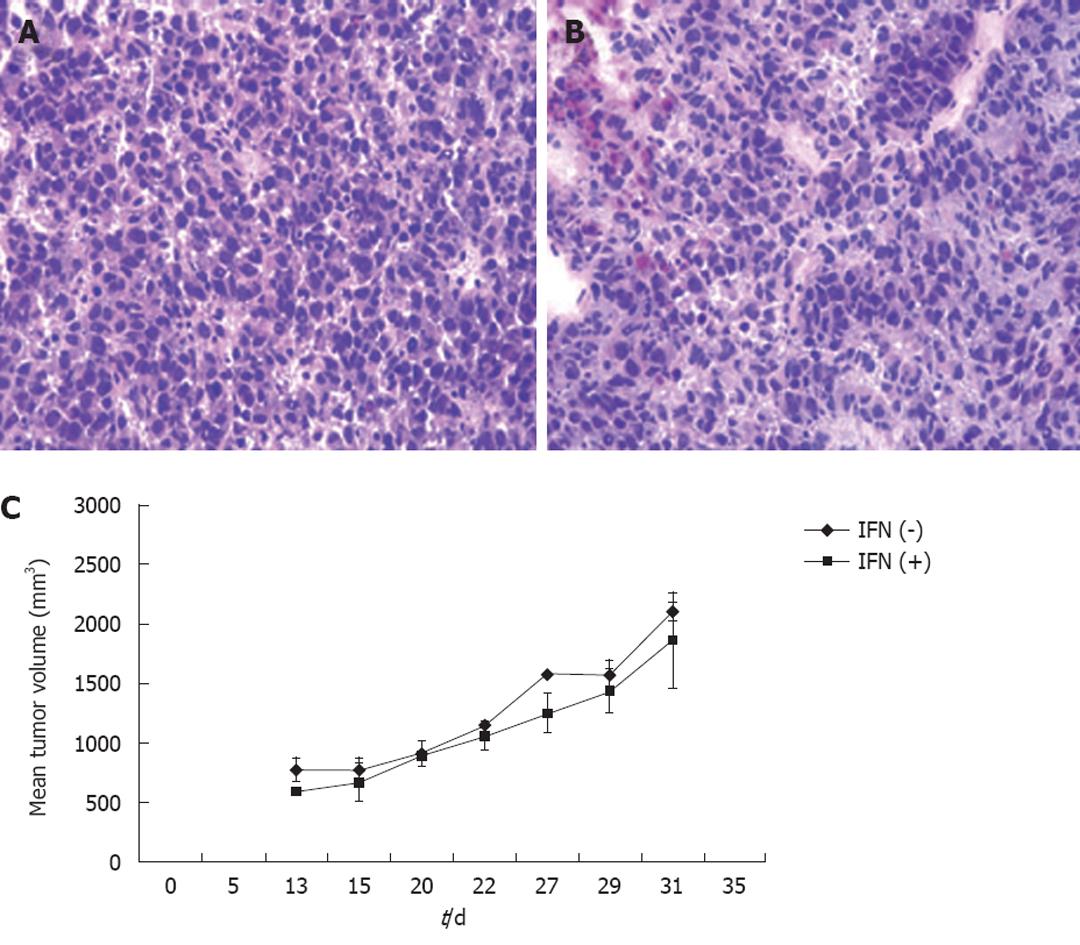
It is important to develop an HCC xenograft in the liver because HCV replicates in the liver of the human host. Furthermore, a liver tumor model is needed to assess and optimize the targeted delivery of antiviral therapy. We wanted to determine whether the mouse-adapted replicon cells that formed a subcutaneous tumor could also form HCC in NOD/SCID mouse liver. The Huh-7 cells were infused into the liver by intrasplenic injection, and examined for liver tumor development at weekly intervals. The growth of mouse-adapted Huh-7 cells in the liver was slow compared to the subcutaneous model, and took almost 4 wk to reveal diffuse HCC nodules throughout the liver (Figure 3C and D). Normal mouse liver and histology are shown in Figure 3A and B. To examine whether HCV replication in liver tumors occurred at a similar level as that seen in the subcutaneous model, frozen sections of the liver tumors were examined for GFP expression. To our surprise, the level of HCV RNA replication was low in the liver tumor, and we could not see a very high level of GFP expression in the HCC xenografts formed in the SCID mouse liver. The loss of GFP expression was not due to an alteration in the open reading frame of GFP in the sub-genomic HCV RNA, because we could recover HCV GFP RNA levels when cells were isolated from the xenografts and cultured in medium supplemented with G-418. The negative GFP expression was due to the low level of HCV replication in the liver tumors. These results suggested that the liver microenvironment significantly suppressed HCV RNA replication compared to that in the subcutaneously formed tumors. We then examined whether these HCV GFP replicon cells needed to adapt to the liver microenvironment to support high-level replication of HCV. The mouse-adapted GFP replicon cells were therefore adapted to the liver microenvironment by another three passages. At each passage, we could recover HCV GFP expression in culture, which suggested that HCV RNA replication in the liver tumors was not completely lost. We then examined whether GFP expression could be seen in the HCC xenografts formed in the mouse liver, using the cells that had undergone three passages. We were able to see GFP expression in tumor nodules in the liver after three passages (Figure 3E and F). The replication of HCV RNA in the liver tumors was also confirmed by cell colony assay after culturing tumor cells in medium that contained G-418 (1 mg/mL). The replicon cells derived from the liver tumors formed G-418-resistant cell clones, which suggested that HCV replication occurred within the HCC xenografts in SCID mice. Liver tumor supporting HCV RNA replication was confirmed by RT-nested PCR followed by Southern blot analysis. These results suggest that we have developed a mouse-adapted S3-GFP replicon cell line that forms liver tumor after splenic injection. The level of HCV RNA in the liver tumor was determined by real time RT-qPCR that was in the range of 104 copies/μg of total cellular RNA. The subcutaneous tumor tissue showed a mean titer of 105 copies/μg of total cellular RNA. There is one log reduction in the HCV RNA titer in the HCC tumor formed subcutaneously vs those formed in the liver (Figure 4). The HCV RNA titer in the liver tumor model is comparable to that found in chronically infected human liver tissue[23]. We hypothesize that this might be due to the fact that there are differences in the innate immune pressure on HCV RNA replication in the peripheral compared with internal organs such as the liver. There is evidence to support our observations that the liver has its own innate immune system that might suppress replication of HCV[24-26].
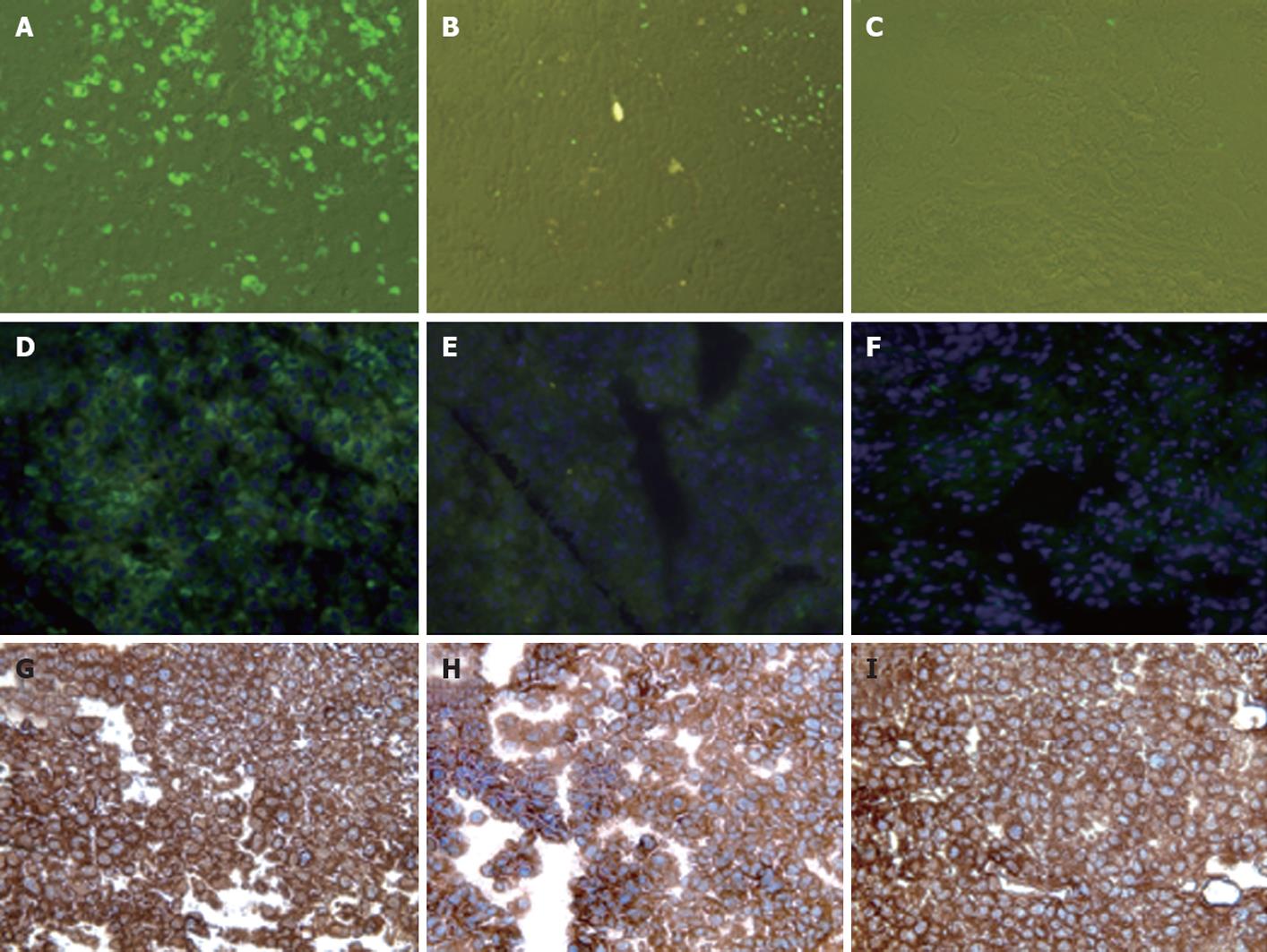
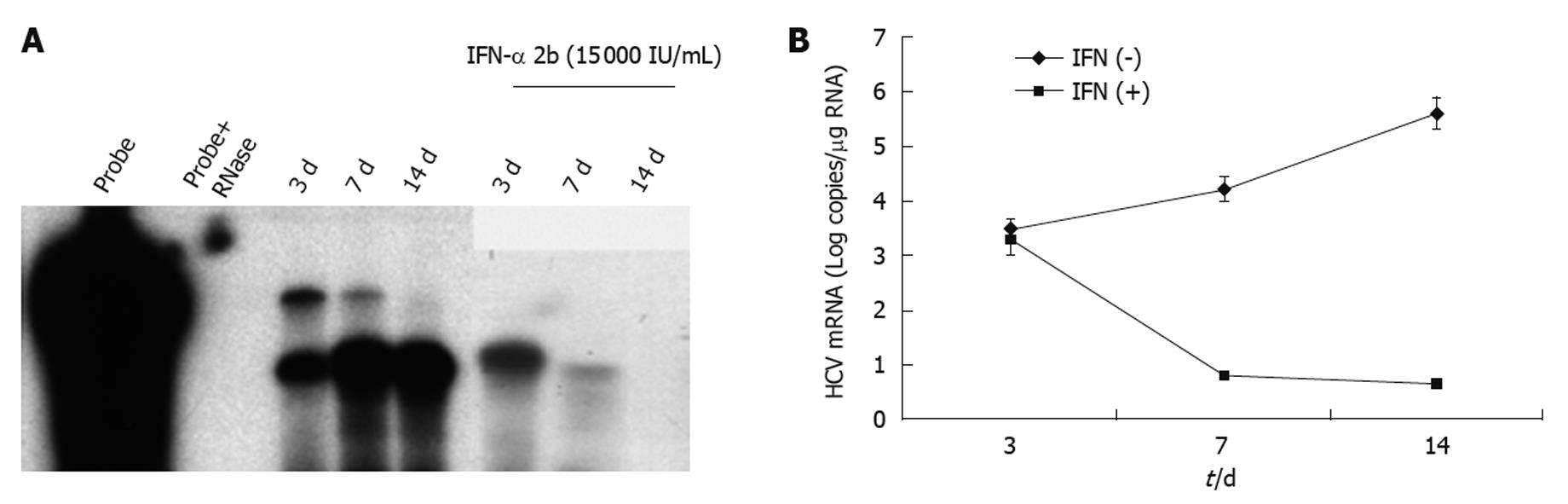
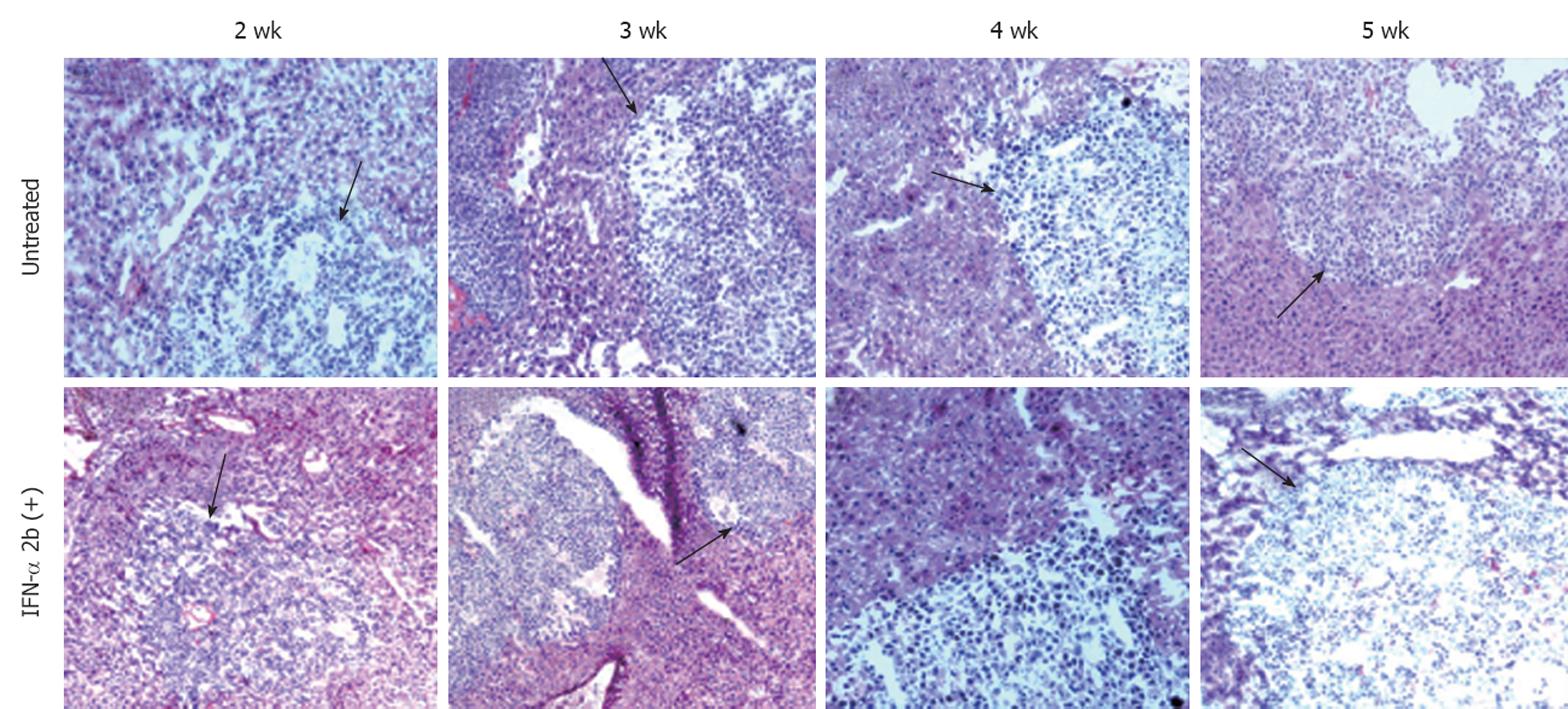
To investigate the ability of IFN-α to inhibit HCV RNA replication in the subcutaneous HCC xenografts, the mice at 2 wk post-implantation were treated with IFN-α at 15 000 IU/mice three times weekly. The mice were sacrificed at different time points after initiation of IFN-α treatment and the tissues were processed for HCV expression. IFN treatment decreased GFP expression in a time-dependent manner, with > 50% decrease in GFP expression after 1 wk (Figure 5B and E), with complete inhibition after 2 wk (Figure 5C and F). The untreated sections showed expression of GFP (Figure 5A and D). To show that the hepatocytes that support HCV replication are human hepatoma cells, immunostaining of HCC cells was performed using an antibody to human serum albumin. The results presented in Figure 5G-I clearly showed that the HCC xenografts were positive for human albumin. The level of albumin expression did not change due to IFN-α treatment. To ensure that IFN-α treatment inhibited viral RNA replication and reduced the level of HCV RNA in the HCC xenografts, total RNA was extracted from the tumor samples and processed for RPA. The results of this analysis are shown in Figure 6, which indicated that HCC xenografts in SCID mice supported a high level of HCV RNA replication, and the levels of HCV RNA in the tumors were completely inhibited after 2 wk of IFN-α treatment. The results shown in Figure 6A and B suggested that HCV RNA level in the subcutaneous HCC tumors increased from 18 to 28 d after tumor development. IFN treatment successfully inhibited virus replication within the tumors and the HCV-RNA titer decreased significantly after 2 wk of treatment, and remained below the limits of detection at 4 wk. We examined the possibility that IFN-α treatment could have an effect on tumor growth, which could have affected the measured HCV RNA levels in the tumors. The tumor size and histology of IFN-α-treated and untreated animals were compared (Figure 7). We did not notice any difference in the tumor growth or histology between the treated and untreated animals. The dose of IFN-α treatment used in the in vivo experiments only inhibited HCV RNA replication, but did not inhibit the tumor growth. Taken together, these results indicated that HCC xenografts that formed subcutaneously in the SCID mice supported a high level of HCV RNA replication, and that IFN-α directly inhibited HCV replication in the tumors. All mice were doing well throughout the IFN-α treatment experiment and their body weight did not change significantly. Mice tolerated the IFN-α treatment well.
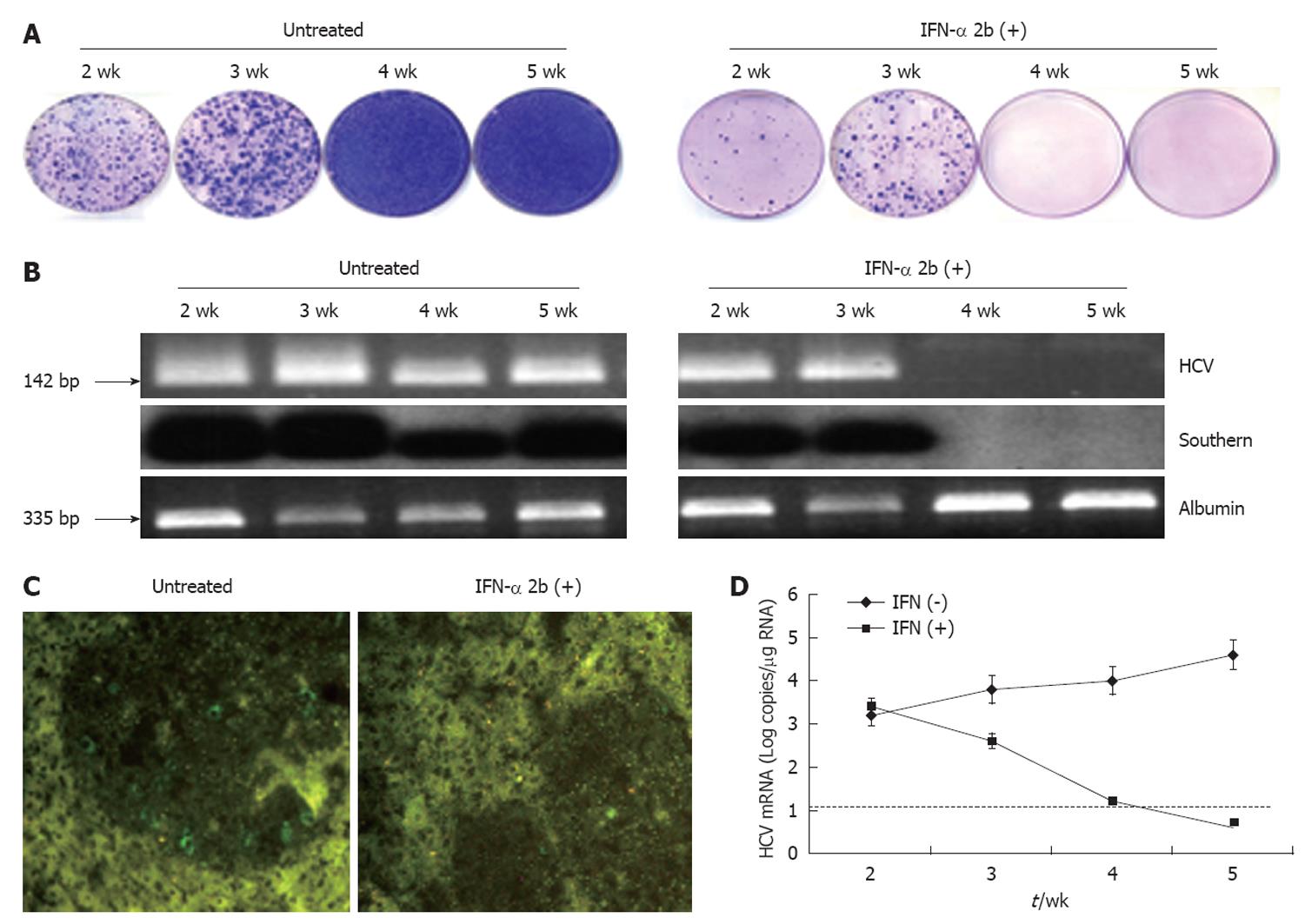
We demonstrated the usefulness of this model by evaluating whether HCV replication in the liver tumor model could be inhibited by IFN-α. Mice were treated with intraperitoneal injection of IFN-α three times weekly at 15 000 IU/mouse, starting at 4 wk after S3-GFP transplantation. Control animals received saline injections at similar time intervals. At different treatment intervals, mice were sacrificed and liver tissue was collected. The ability of IFN-α to inhibit HCV replication was confirmed by four independent assays. First, hepatocytes were isolated from the liver and cultured in the presence of growth medium that contained G-418 (1 mg/mL). The number of S3-GFP replicon cell colonies in the plate was compared to assess the antiviral effects of IFN-α against HCV replication in the xenograft tumors. Figure 8A shows that 4 wk IFN-α treatment successfully inhibited replication of HCV in the liver tumors. Second, the intracellular HCV RNA in the tumors tissue was examined by using RT-nested PCR and Southern blot analysis that targeted the highly conserved 5’-UTR region of the HCV genome. We showed that HCV RNA was detectable in the liver tumors of untreated mice over 5 wk, by RT-nested PCR and Southern blot analysis. IFN-α treatment completely inhibited HCV RNA replication in the liver tumors and remained undetectable at 4 and 5 wk, using the highly sensitive RT-nested PCR and Southern blot analysis (Figure 8B). IFN-α treatment did not alter the cellular albumin RNA levels. Third, expression of GFP in the liver tumors after IFN-α treatment was negative after 4 wk (Figure 8C). Fourth, the level of HCV RNA in the treated and untreated tumor samples was assessed by real-time PCR. The results of this analysis suggested that HCV levels decreased significantly in the tumors and remained below the detection limit after 4 wk of IFN-α treatment (Figure 8D). These data clearly showed that IFN-α inhibited HCV RNA replication in the liver tumor model and there was a significant difference in the level of HCV replication in the liver tumors between the untreated and IFN-α-treated mice. To demonstrate that the decrease in the HCV RNA replication after IFN-α treatment was not due to the direct effect of IFN on tumor growth in these HCC xenografts, histological examination of liver tumor in IFN-α-treated and untreated animals was performed. The histological pictures shown in Figure 9 indicate that there was no evidence of cellular necrosis in the tissue sections of mice that received IFN-α treatment. These results indicated that the reduction of HCV RNA levels in the tumor was due to a specific antiviral mechanism of IFN-α.
Cell culture and animal models are required for the development of novel experimental therapies for chronic HCV infection. A number of antiviral strategies have been developed that inhibit HCV replication in cell culture systems[4,5]. The successful use of different therapeutic agents to inhibit chronic HCV infection requires further validation using an easily accessible small animal model. Our laboratory has developed multiple siRNA targets in the 5’-UTR region of the HCV genome, and has shown that intracellular delivery of these siRNAs can completely degrade HCV RNA in cell culture[27]. We have also developed recombinant antibodies targeted to the viral NS3 helicase, and have shown that intracellular expression of these antibodies inhibits viral helicase activity and virus replication in a cell culture model[28]. The next step of this research is to test the effectiveness of these different antiviral strategies that target inhibition of viral replication, using an animal model system. Therefore, development of a small animal model for HCV infection was essential to optimize targeted, hepatic delivery of siRNA and recombinant antibodies. Mice and other rodents are not susceptible to natural HCV infection, thus, there are several alternative approaches that have been employed during the past few years to replicate HCV infection using mouse models. The advantages and disadvantages of these models to study HCV infection in mouse models are discussed.
The most promising small animal model for studying HCV infection is now the human liver chimeric mouse. The principle of developing this mouse model is based on the availability of specific mouse strains in which human hepatocytes can be efficiently transplanted into the mouse liver. Two specific mouse strains are currently being used for development of the humanized mouse liver. One is the SCID-Alb/uPA mouse and the other is the Fah-/- Rag-/- IL-2-/- mouse. Both mouse strains have high liver regeneration capacity due to their liver injury and support a high degree of human hepatocyte transplantation. The humanized mouse liver has been shown to have normal hepatic architecture and support HCV replication after natural infection. The studies conducted by Mercer et al[13] and Tateno et al[29] have used a mouse strain called SCID-Alb/uPA. Using this mouse strain, Mercer et al[13] have shown that the humanized mouse liver contains > 50% human hepatocytes, and Tateno et al[29] have shown that humanized mouse liver contains > 92% human hepatocytes. The Alb/uPA transgenic mouse strain shows severe hepatotoxicity due to overexpression of the urokinase plasminogen (uPA) gene in hepatocytes, which leads to continuous liver regeneration. Human hepatocytes that lack this transgene preferentially survive after transplantation and are stably maintained in this mouse liver. This mouse strain has been crossed with an immunodeficient mouse (SCID or Rag2) to generate the SCID-Alb/uPA mouse strain that tolerates transplantation of human hepatocytes well. Human hepatocytes can be successfully transplanted into the SCID-Alb/uPA mice within 1 or 2 wk after birth. Several studies have now successfully used this model to replicate HCV infection and have tested several antiviral molecules, including IFN-α and protease inhibitors. The study conducted by Bissig et al[16] has used Fah-/- Rag2-/- IL-2-/- mice and has shown that the humanized mouse liver contains > 80% human hepatocytes. This mouse strain lacks fumaryl acetoacetate, which is an enzyme that is required for tyrosine amino acid metabolism. Mice that lack this enzyme show severe hepatotoxicity due to accumulation of toxic metabolites. This mouse strain has been bred with Rag2-/- IL-2-/- mice to generate an immunodeficient mouse strain that supports transplantation of adult human hepatocytes. Recently, Bissig et al[16] have used this mouse model to replicate HCV infection from cell culture and from the serum of chronically infected patients. They also have shown that pegylated IFN-α and Debio 025 (cyclophilin inhibitor) inhibit HCV replication in this chimeric mouse model. The investigators have shown that these chimeric mice sustain viral replication for > 6 mo, without any substantial side effects. There is no doubt that these chimeric mouse models are extremely challenging technically, complicated to use, and expensive. Despite the technical challenges, these models hold promise for studying the long-term effect of HCV infection on the development of liver fibrosis and cancer.
We described here an HCC xenograft mouse model in which human tumor cells were implanted in the liver of an immunodeficient mouse. This model is relatively simple compared to the human liver chimeric mouse model. This model can be used for experimental verification of the efficacy of targeted antiviral therapies against HCV. The model utilizes SCID/bg and NOD/SCID mice with a partially suppressed innate immune system (natural killer cell response) and adaptive immune system (both B- and T-cell mediated response)[30]. In an earlier study, Zhu et al[18] have performed similar experiments to generate a mouse-adapted HCV Con-1 replicon cell line using the SCID/bg mice. The in vivo adaptation of HCV is not possible using this approach due to the low-level replication capacity of the HCV Con-1 replicon clone. The authors had to re-engineer the sub-genomic clone with reinsertion of HCV sub-genomic RNA with a highly efficient internal ribosome entry site, and a highly adapted mutation in the non-structural gene, to sustain high-level replication of HCV RNA within the tumor. They have successfully measured HCV replication in this mouse model by measuring luciferase activity using non-invasive whole-body imaging. They have shown that IFN-α and protease inhibitor BILN-2061 reduce HCV RNA replication in the subcutaneous and liver tumor models. The authors have not shown whether these treatments lead to complete clearance of HCV in these tumors models. Another study by Guévin et al[31] also has demonstrated that full-length HCV replication can be achieved in HCC tumor xenografts formed subcutaneously using immunodeficient mouse strains. The authors have made mouse-adapted Huh-7 cells by multiple passages in SCID mice. The authors have shown that the xenograft tumors in the SCID mouse produce infectious virus particles. The infectious virus produced in the xenograft tumors can be serially passaged in SCID mice. The authors have shown that IFN-α and protease inhibitor BILN-2061 inhibit replication of the cell culture HCV cc strain JFH1 in the mouse model.
The mouse model described here differs in some aspects compared to the two previously described tumor xenograft models. We realized that high-level replication of HCV in the SCID mouse liver might be possible by using the highly efficient JFH1 clone. We used the JFH1 virus clone that replicated at a much higher level in the HCC xenografts formed in the SCID mice. The GFP-labeled replicon cell line showed that replication of HCV occurred in the HCC tumors, and could be determined by direct examination under fluorescence microscopy. The replicon cell line is similar to chronically infected human hepatocytes, except that these cells do not produce the infectious virus, which makes this animal model safer to use. We have used a number of biochemical methods (colony assay, RPA, real-time RT-PCR and RT-nested PCR and Southern blot analysis) that quantitatively evaluate HCV replication in the subcutaneous tumors in an accurate manner. If necessary, this model can also be adapted using a luciferase-based, mouse-adapted HCV replicon cell line so that the whole-body imaging technique can be used to monitor HCV replication in the mouse model. The diffuse metastasis of HCC throughout the liver lobes after intrasplenic infusion of replicon cells mimics in vivo distribution of chronically infected hepatocytes in human liver. We demonstrated replication of HCV RNA in multiple HCC nodules in the portal tracts of the SCID mice livers. This liver tumor model developed by intrasplenic injection of replicon cells provides a better assessment of the biodistribution and pharmacokinetics of IFN-α when compared to direct injection of replicon cells to the liver, where HCC tumors are formed only in selected areas of the organ. We noticed that the level of HCV replication in the liver tumor model was 10-fold lower when compared to that in the subcutaneous model. This could be due to the innate antiviral pressure against HCV between the peripheral organ and liver microenvironment. The dynamics of viral RNA replication in the liver tumor model are comparable to those of chronically infected human liver. As a practical validation of this mouse model, we showed that IFN-α treatment inhibited HCV RNA replication and abolished GFP expression in the subcutaneous HCC xenografts within 2 wk, which suggests that the mouse model can be used to test the success of other antiviral strategies against HCV. We showed that IFN-α completely inhibited viral replication in the liver tumor model, and HCV RNA in liver tumors was undetectable by using a highly sensitive RT-nested PCR. The sensitivity of this RT-nested PCR assay was found to be in the range of 1-10 copies of HCV RNA[32]. Using this mouse model, we showed that IFN-α administration completely eliminated HCV sub-genomic RNA replication, which supports the practical use of this animal model to test other antiviral strategies against the background of a growing tumor. This mouse model, therefore, can be used practically to optimize methods for targeted delivery of siRNA and recombinant antibodies that are designed to inhibit HCV replication in tumor xenografts in the mouse liver.
The SCID mouse model described here can be adapted to any research laboratory because it is relatively simple, safe and less expensive, thus allowing it to be used in large numbers for testing. This mouse model can be utilized to establish the success of targeted liver delivery methods of a variety of antiviral strategies within a short period of time. In vivo experiments that address the toxicity of different antiviral approaches with this animal model can be tested using a large number of mice to achieve statistical power of significance. This model deals with non-infectious HCV and does not depend on the use of donated or harvested human hepatocytes or infected serum samples. We have also developed in vivo adapted Huh-7 cells by elimination of HCV replicon replication. Tumor xenografts that use this Huh-7 cell line in SCID mice can be used to study the infectivity of JFH1 virus and other relevant clinical HCV strains. This will allow us to test the efficacy of neutralizing monoclonal antibodies directed against the HCV envelope protein. There are also some limitations in the tumor xenograft mouse models for HCV that are described here. For example, this model cannot be used for investigation of immunopathogenesis of chronic HCV infection or vaccine development because the SCID mice have defective B- and T-cell responses. Nevertheless, this animal model will be useful for antiviral evaluation and testing the potential of intracellular delivery of siRNA and recombinant antibodies that are directed at inhibition of HCV replication in the liver.
Hepatitis C virus (HCV) is the most common blood-borne infection that affects the liver. Several antiviral strategies have been developed during the past 2 years, including small molecules, antisense oligonucleotides, siRNA, ribozymes, and recombinant antibodies that are designed to inhibit HCV replication in cell culture models. A small animal model for HCV is required to test different experimental therapies developed using HCV cell culture systems.
Development of a small animal model for studying HCV infection has been difficult because the virus only infects humans and chimpanzees. The only available animal model is a chimeric humanized mouse model. This model is very costly, technically very challenging, and very expensive. The authors developed an hepatocellular carcinoma (HCC) xenograft tumor model in SCID mice using a replicon cell line that replicates highly efficient JFH1 virus. The replicon cells were transfected with a green fluorescent protein (GFP)-tagged HCV sub-genomic clone so that the antiviral effect could be observed by GFP expression. The replication of HCV could be inhibited by interferon-α (IFN-α), which indicates that the model can be used for antiviral screening. The HCC xenograft mouse model is less expensive, non-infectious, and can be easily adapted to any research laboratory.
The authors developed an S3-GFP replicon Huh-7 cell line that was highly adapted to mice and formed human HCC xenografts in SCID mice. We showed that HCV replicated in the HCC xenografts formed subcutaneously and in the liver. The antiviral effect of IFN-α showed clearance of HCV in this mouse model.
This model can be used to optimize methods for liver-targeted delivery of several antiviral strategies against HCV replication.
The paper is well written and the scientific content is worth publishing.
Peer reviewers: Naoaki Sakata, MD, PhD, Division of Hepato-Biliary Pancreatic Surgery, Tohoku University Graduate School of Medicine, 1-1 Seiryo-machi, Aoba-ku, Sendai, Miyagi 980-8574, Japan; Dr. Sara Lindén, PhD, Professor, Mucosal Immunobiology and Vaccine Center, Gothenburg University, Box 435, Göteborg, 405 30, Sweden
S- Editor Cheng JX L- Editor Kerr C E- Editor Zheng XM
| 1. | Williams R. Global challenges in liver disease. Hepatology. 2006;44:521-526. |
| 3. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. |
| 4. | Webster DP, Klenerman P, Collier J, Jeffery KJ. Development of novel treatments for hepatitis C. Lancet Infect Dis. 2009;9:108-117. |
| 5. | Pawlotsky JM, Chevaliez S, McHutchison JG. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology. 2007;132:1979-1998. |
| 6. | Alter HJ, Purcell RH, Holland PV, Popper H. Transmissible agent in non-A, non-B hepatitis. Lancet. 1978;1:459-463. |
| 7. | Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606-614. |
| 8. | Fournier C, Sureau C, Coste J, Ducos J, Pageaux G, Larrey D, Domergue J, Maurel P. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J Gen Virol. 1998;79:2367-2374. |
| 9. | Kremsdorf D, Brezillon N. New animal models for hepatitis C viral infection and pathogenesis studies. World J Gastroenterol. 2007;13:2427-2435. |
| 10. | Barth H, Robinet E, Liang TJ, Baumert TF. Mouse models for the study of HCV infection and virus-host interactions. J Hepatol. 2008;49:134-142. |
| 11. | Wu GY, Konishi M, Walton CM, Olive D, Hayashi K, Wu CH. A novel immunocompetent rat model of HCV infection and hepatitis. Gastroenterology. 2005;128:1416-1423. |
| 12. | Ilan E, Arazi J, Nussbaum O, Zauberman A, Eren R, Lubin I, Neville L, Ben-Moshe O, Kischitzky A, Litchi A. The hepatitis C virus (HCV)-Trimera mouse: a model for evaluation of agents against HCV. J Infect Dis. 2002;185:153-161. |
| 13. | Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927-933. |
| 14. | Kneteman NM, Weiner AJ, O'Connell J, Collett M, Gao T, Aukerman L, Kovelsky R, Ni ZJ, Zhu Q, Hashash A. Anti-HCV therapies in chimeric scid-Alb/uPA mice parallel outcomes in human clinical application. Hepatology. 2006;43:1346-1353. |
| 15. | Turrini P, Sasso R, Germoni S, Marcucci I, Celluci A, Di Marco A, Marra E, Paonessa G, Eutropi A, Laufer R. Development of humanized mice for the study of hepatitis C virus infection. Transplant Proc. 2006;38:1181-1184. |
| 16. | Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924-930. |
| 17. | Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Res. 2008;80:231-238. |
| 18. | Zhu Q, Oei Y, Mendel DB, Garrett EN, Patawaran MB, Hollenbach PW, Aukerman SL, Weiner AJ. Novel robust hepatitis C virus mouse efficacy model. Antimicrob Agents Chemother. 2006;50:3260-3268. |
| 19. | Hazari S, Chandra PK, Poat B, Datta S, Garry RF, Foster TP, Kousoulas G, Wakita T, Dash S. Impaired antiviral activity of interferon alpha against hepatitis C virus 2a in Huh-7 cells with a defective Jak-Stat pathway. Virol J. 2010;7:36. |
| 20. | Ponder KP, Gupta S, Leland F, Darlington G, Finegold M, DeMayo J, Ledley FD, Chowdhury JR, Woo SL. Mouse hepatocytes migrate to liver parenchyma and function indefinitely after intrasplenic transplantation. Proc Natl Acad Sci USA. 1991;88:1217-1221. |
| 21. | Akyol G, Dash S, Shieh YS, Malter JS, Gerber MA. Detection of hepatitis C virus RNA sequences by polymerase chain reaction in fixed liver tissue. Mod Pathol. 1992;5:501-504. |
| 22. | Gibellini D, Gardini F, Vitone F, Schiavone P, Furlini G, Re MC. Simultaneous detection of HCV and HIV-1 by SYBR Green real time multiplex RT-PCR technique in plasma samples. Mol Cell Probes. 2006;20:223-229. |
| 23. | Bellecave P, Sarasin-Filipowicz M, Donzé O, Kennel A, Gouttenoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology. 2010;51:1127-1136. |
| 24. | Swenson ES, Price JG, Brazelton T, Krause DS. Limitations of green fluorescent protein as a cell lineage marker. Stem Cells. 2007;25:2593-2600. |
| 25. | Exley MA, Koziel MJ. To be or not to be NKT: natural killer T cells in the liver. Hepatology. 2004;40:1033-1040. |
| 26. | Nemeth E, Baird AW, O'Farrelly C. Microanatomy of the liver immune system. Semin Immunopathol. 2009;31:333-343. |
| 27. | Prabhu R, Garry RF, Dash S. Small interfering RNA targeted to stem-loop II of the 5' untranslated region effectively inhibits expression of six HCV genotypes. Virol J. 2006;3:100. |
| 28. | Prabhu R, Khalap N, Burioni R, Clementi M, Garry RF, Dash S. Inhibition of hepatitis C virus nonstructural protein, helicase activity, and viral replication by a recombinant human antibody clone. Am J Pathol. 2004;165:1163-1173. |
| 29. | Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, Soeno Y, Asahina K, Hino H. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901-912. |
| 30. | Shibata S, Asano T, Ogura A, Hashimoto N, Hayakawa J, Uetsuka K, Nakayama H, Doi K. SCID-bg mice as xenograft recipients. Lab Anim. 1997;31:163-168. |
| 31. | Guévin C, Lamarre A, Labonté P. Novel HCV replication mouse model using human hepatocellular carcinoma xenografts. Antiviral Res. 2009;84:14-22. |
| 32. | Dash S, Halim AB, Tsuji H, Hiramatsu N, Gerber MA. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am J Pathol. 1997;151:363-373. |