Published online Nov 28, 2010. doi: 10.3748/wjg.v16.i44.5582
Revised: August 15, 2010
Accepted: August 23, 2010
Published online: November 28, 2010
AIM: To develop a sensitive assay for screening compounds against hepatitis C virus (HCV).
METHODS: The proteolytic cleavage of NS3/4A on enhanced yellow fluorescent protein (eYFP)-mitochondrial antiviral signaling protein (MAVS) was examined by reporter enzyme secreted placental alkaline phosphatase (SEAP), which enabled us to perform ongoing monitoring of anti-HCV drugs through repeated chemiluminescence. Subcellular localization of eYFP-MAVS was assessed by fluorescence microscopy. Cellular localization and protein levels were examined by Western blotting.
RESULTS: HCV NS3/4A protease cleaved eYFP-MAVS from mitochondria to block the activation of interferon (IFN)-β promoter, thus resulting in downregulation of SEAP activity. The decrease in SEAP activity was proportional to the dose of active NS3/4A protease. Also this reporter assay was used to detect anti-HCV activity of IFN-α and cyclosporine A.
CONCLUSION: Our data show that this reporter system is a sensitive and quantitative reporter of anti-HCV inhibitors. This system will constitute a new tool to allow the efficient screening of HCV inhibitors.
- Citation: Fu QX, Wang LC, Jia SZ, Gao B, Zhou Y, Du J, Wang YL, Wang XH, Peng JC, Zhan LS. Screening compounds against HCV based on MAVS/IFN-β pathway in a replicon model. World J Gastroenterol 2010; 16(44): 5582-5587
- URL: https://www.wjgnet.com/1007-9327/full/v16/i44/5582.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i44.5582
Hepatitis C virus (HCV) infection represents a major public health threat because of the chronic nature of the infection, its high prevalence and the significant morbidity of the resultant disease[1-3]. There is no vaccine, and drug treatment is costly and has poor efficacy[4,5]. New treatment regimens that are more efficacious and better tolerated by all patients are needed. Many new antiviral compounds have been synthesized; several of which have shown promising results in early clinical trials[6,7]. However, development of new drugs is hampered by the lack of appropriate model systems. Therefore, there is an obvious and urgent need to develop a more effective and sensitive assay to facilitate the search for improved antivirals.
HCV is an enveloped virus that contains a single-stranded, positive-sense RNA genome of approximate 9.6 kb. The HCV genome encodes a large open reading frame that is flanked by structured 5’ and 3’ non-translated regions. There are at least 10 mature proteins that are contained in the polyprotein in the order: NH2-core/E1/E2/p7/NS2/NS3/NS4A/NS4B/NS5A/NS5B-COOH, which is post-translationally processed. Processing of the nonstructural part of the polyprotein is mediated by the nonstructural NS3/4A protease and helicase[8-10]. Also, NS3/4A blocks the cellular interferon (IFN) response to double-stranded RNA by proteolytic cleavage of mitochondrial antiviral signaling protein (MAVS; also known as IPS-1, VISA, and CARDIF) or of the adaptor molecule Toll/interleukin-1 receptor domain-containing adaptor inducing IFN-β (TRIF)[11-16].
The ability of NS3/4A protease to control the antiviral host response to HCV facilitated our study to develop a new assay for drug screening. Here, we extended the utility of enhanced yellow fluorescent protein (eYFP)-MAVS/IFN-β-secreted placental alkaline phosphatase (SEAP) signaling pathway for anti-HCV drug discovery. Expression of NS3/4A protease in replicon cells disrupted the eYFP-MAVS/IFN-β-SEAP signaling pathway by proteolytic cleavage of eYFP-MAVS, which resulted in the loss of mitochondrial localization and abrogation of the eYFP-MAVS/IFN-β-SEAP signaling pathway. The decrease in SEAP activity was proportional to the dose of active NS3/4A protease. This reporter assay also was used to detect the activity of IFN-α and cyclosporine A (CsA). Our data show that this reporter system is a sensitive and quantitative reporter assay for anti-HCV inhibitors. This system will constitute a new tool to allow the efficient screening of HCV inhibitors.
Plasmids eYFP-MAVS and pCon1-FL were kindly provided by Martin Baril and Dr. Charles M. Rice of Rockefeller University, respectively. Reporter plasmid of IFN-β-SEAP controlled by the promoter region of IFN-β (from -281 to 20) was constructed based on pTAL-SEAP (Clontech, Mountain View, CA, USA). The NS3/4A sequence derived from HCV type 1b (pCon1-FL) was cloned into pCI Mammalian Expression Vector using EcoRI and SalI sites.
Huh-7.5 and HCV replicon cells that contained full-length HCV sequences of genotype 1b were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Biochrom AG, Berlin, Germany) and penicillin (100 μg/mL)/streptomycin (100 μg/mL). In 24-well plates, cells were seeded at a density of 6 × 104 cells per well in 500 μL DMEM/10% FBS. After incubation at 37°C overnight, or until cells were approximately 70% confluent, cells were transfected with various plasmid constructs using Lipofectamine™ 2000 reagent (Invitrogen, Carlsbad, CA, USA). As an internal control for transfection efficiency, 0.02 μg Renilla luciferase plasmid, pRL-TK (Promega, Madison, WI, USA) was co-transfected with 1 μg of each expression vector in each experiment.
Huh-7.5 and HCV replicon cells were seeded in 24-well plates at a density of 2 × 104 cells per well. After incubation at 37°C overnight, cells were treated with various concentrations of IFN-α (Schering-Plough, Kenilworth, NJ, USA) or CsA (Sigma, St Louis, MO, USA). Two or three days later, cells were transfected with eYFP-MAVS and IFN-β-SEAP with the same concentration of drugs, and cells were incubated for one or two more days. Cell culture was collected and SEAP activity was measured by chemiluminescence. NS3 expression level in replicon cells was determined by Western blotting.
SEAP activity was measured using the Phospha-Light™ assay kit (Tropix, Foster City, CA, USA) according to the manufacturer’s instructions.
Protein concentration was estimated by BCA™ Protein Assay Kit from Pierce (Rockford, IL, USA). Cell lysates were separated by SDS-PAGE, and protein expression was detected by Western blotting analysis using antibodies. eYFP was detected using a rabbit polyclonal antibody (Proteintech, Chicago, IL, USA), actin was detected using a rabbit polyclonal antibody (Sigma), NS3 was detected using a goat polyclonal antibody (LifeSpan BioSciences, Seattle, WA, USA), with horseradish peroxidase (HRP)-conjugated secondary antibody followed by detection by chemiluminescent HRP substrate (Millipore, Billerica, MA, USA).
All results were expressed as the mean ± SD. Statistical comparisons between two groups were made using Student’s t test after analysis of variance. The level of significance was set at α = 0.05.
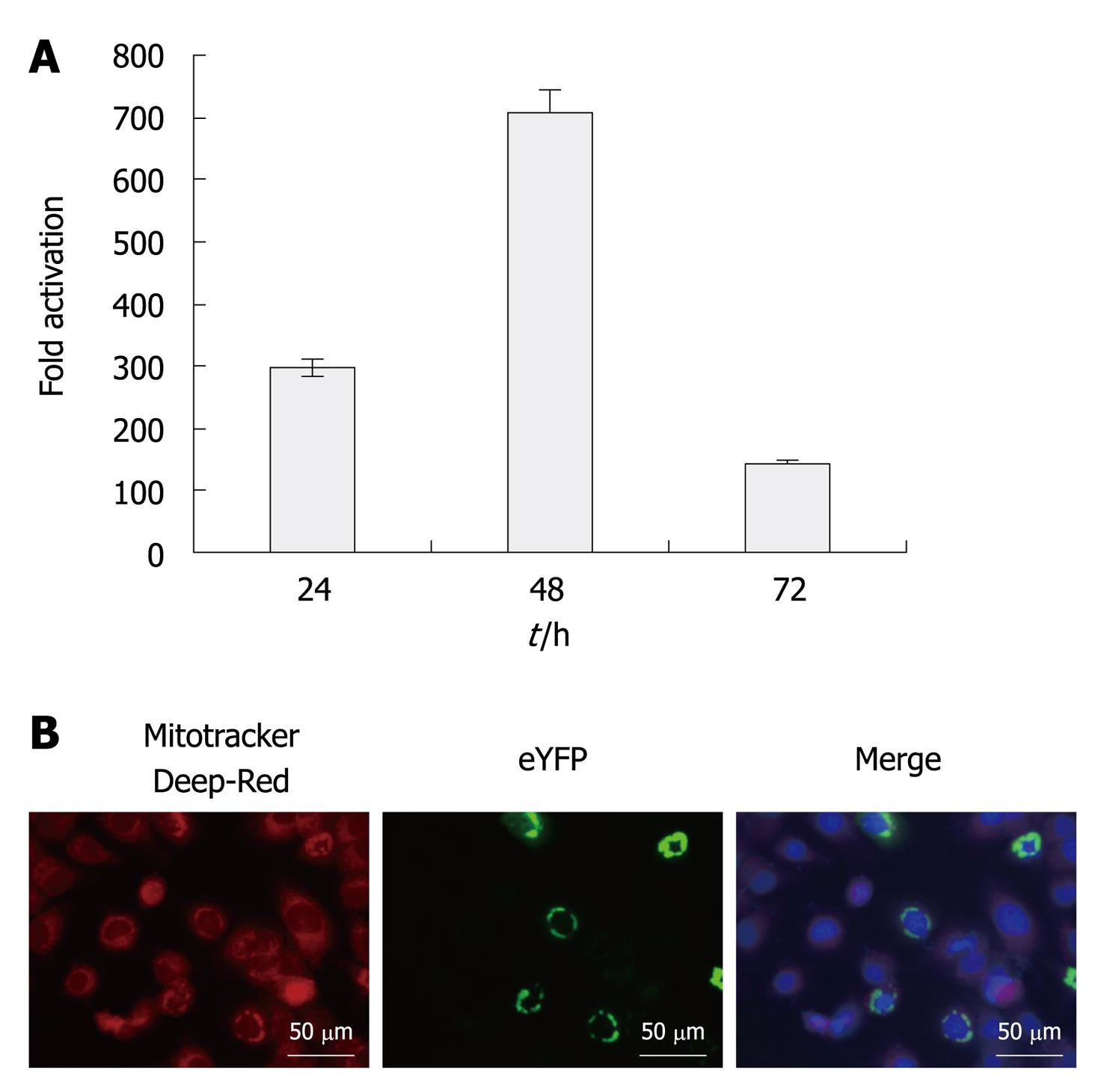
We assessed signaling activity of eYFP-MAVS in Huh7.5 cells by co-transfection alongside IFN-β-SEAP. As expected, eYFP-MAVS induced the activation of the IFN-β promoter. As shown in Figure 1, at 48 h post-transfection, eYFP-MAVS gave rise to an approximately 700-fold increase in SEAP activity. Subcellular localization of eYFP-MAVS was also assessed by fluorescence microscopy, with cells expressing eYFP-MAVS proteins. Prior to visualization, mitochondria and nuclei were labeled with Mitotracker deep red and 4’,6-Diamidino-2-phenylindole (DAPI), respectively. Figure 1 shows eYFP-MAVS localized to the mitochondrial membrane.
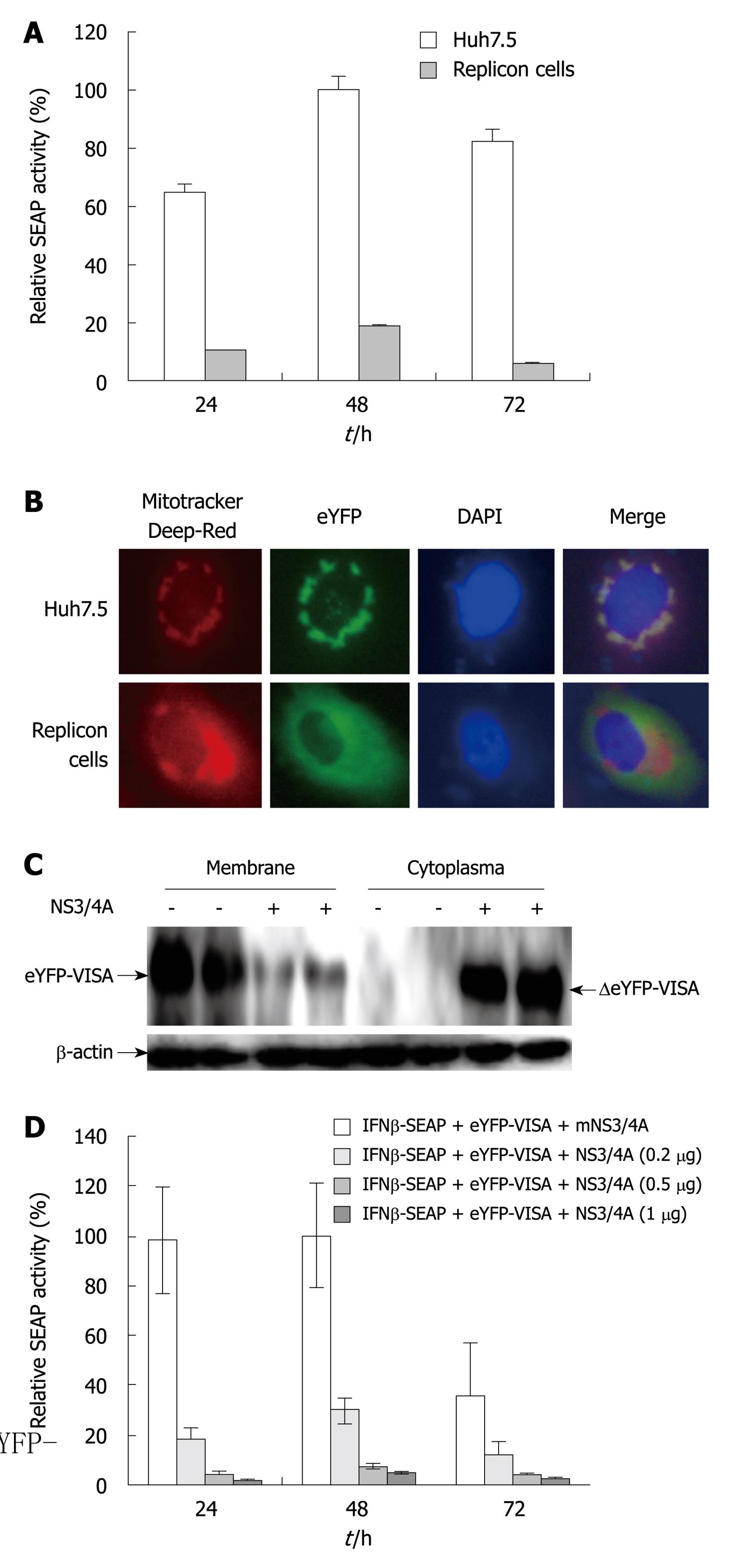
Our assay was used to assess HCV replication in Huh7.5 cells that stably expressed full-length HCV replicons. The replicon cell lines were co-transfected with eYFP-MAVS and IFN-β-SEAP, and naive Huh7.5 cells were simultaneously transfected to serve as a control. SEAP activity in HCV replicon cells was approximately 20% relative to that in the control group (P < 0.05, Figure 2A). In the presence of HCV NS3/4A protease, eYFP-MAVS was proteolytically cleavaged as reported previously[11,16]. The proteolytically cleaved eYFP-MAVS, named ΔeYFP-MAVS, only could be detected in HCV replicon cells (Figure 2C), whose localization shifted from the mitochondrial membrane to the cytoplasm (Figure 2B).
The sensitivity of this assay was examined by co-transfecting eYFP-MAVS and IFN-β-SEAP with various concentrations of pNS3/4A, or with the control empty vector. SEAP activity was evaluated 24, 48 and 72 h post-transfection. The expression of NS3/4A protease in transfected cells resulted in the expected downregulation of the eYFP-MAVS/IFN-β-SEAP signaling pathway in a dose-dependent manner (P < 0.05, Figure 2D). These results indicated that this reporter system could be used for quantitative analysis of NS3/4A protease activity.
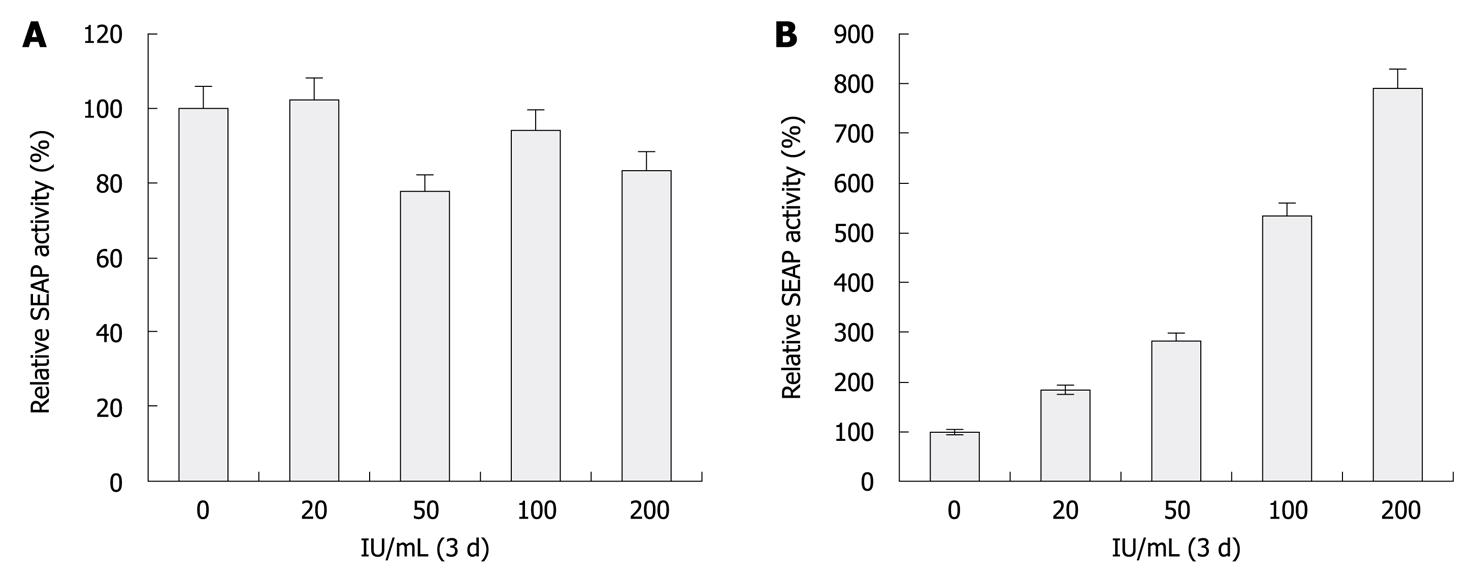
To show the feasibility of this reporter system for characterizing HCV inhibitors, Huh-7.5 and HCV replicon cells incubated with various concentrations of IFN-α, ranging from 0 to 200 U/mL for 72 h were transfected with eYFP-MAVS and IFN-β-SEAP. At 48 h, SEAP activity of replicon cells was proportional to the extent of anti-HCV treatment (P < 0.05, Figure 3A). Meanwhile, Western blotting analysis of these samples revealed the amount of NS3 protease that was encoded by the HCV replicon was proportionally reduced by treatment with increasing concentrations of IFN-α (Figure 3B). The increase in SEAP activity was inversely proportional to the dose of active NS3/4A protease. No difference was observed with Huh-7.5 cells. These results indicate that this reporter system could be used for quantitative analysis of HCV-inhibitory compounds in cell culture systems.
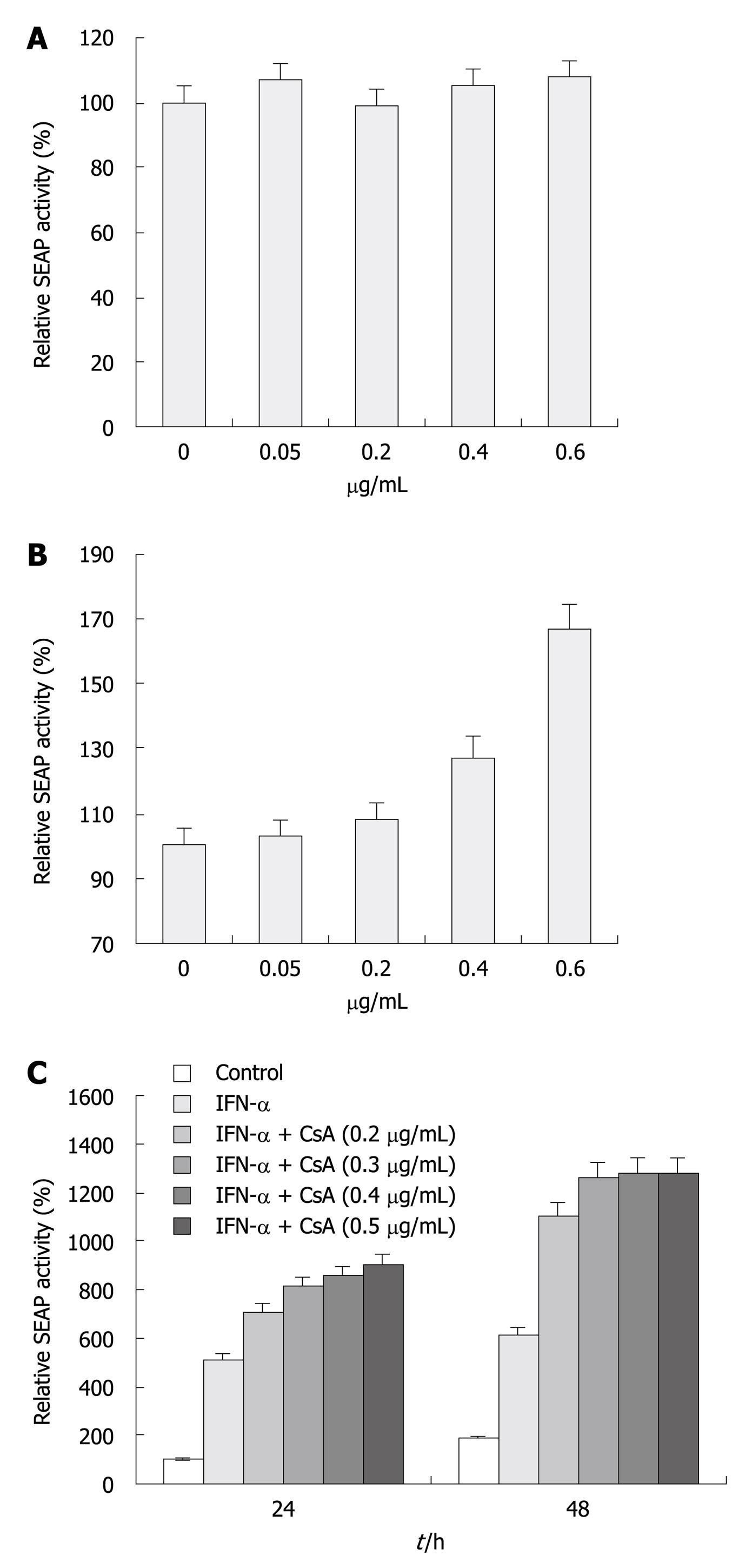
CsA is a well-characterized anti-HCV reagent that strongly suppresses viral replication in cell culture[17-19]. We evaluated the inhibitory effect of CsA on HCV replication and its combination effect with IFN-α. Huh-7.5 and replicon cells treated with increasing doses of CsA (Figure 4A) for 48 h were transfected with eYFP-MAVS and IFN-β-SEAP. At 24 h post-transfection, cells treated with 0.4 and 0.6 μg/mL CsA had significantly higher SEAP activity compared with cells treated with vehicle or 0.05 and 0.2 μg/mL CsA, whereas no difference was observed with Huh-7.5 cells (Figure 4A). As shown in Figure 4B, IFN-α in combination with CsA therapy enhanced its inhibitory effect. Cells treated with both agents had significant enhancement in SEAP activity compared with cells treated with a single agent. Determination of SEAP activity revealed that 0.2 μg/mL CsA-treated cells had a minimal increase in SEAP activity compared to untreated cells (Figure 4A), whereas, in combination with IFN-α, 0.2 μg/mL CsA-treated cells had a 5-6-fold increase. Cells treated with CsA and IFN-α showed a > 7-fold increase in SEAP activity.
To date, the only available therapy for chronic hepatitis C is IFN-α, either alone or in combination with the nucleoside analogue ribavirin. However, many patients are discouraged from IFN-based treatment because of severe side effects. In the past few years, much progress has been made in the development of small-molecule-based therapeutics for the treatment of HCV infection, such as inhibitors of HCV enzymes, as well as nucleic-acid-based agents that attack the viral RNA. However, the lack of appropriate model systems for drug evaluation has hampered their clinical use.
Although the replicon systems and the infectious HCV cell culture system have served as extremely valuable tools for in vitro study of HCV replication, and for screening and evaluation of new antiviral drugs against HCV that specifically target the protease activity of NS3 or the polymerase activity of NS5[20-26], the level of viral multiplication is not satisfactory, because in all cases, the detection of viral RNA and protein has to rely on reverse transcriptase-polymerase chain reaction and antibody labeling, which have some inherent technical problems that are difficult to overcome. To date, many different HCV replicons have been generated with other reporter genes such as firefly luciferase or fluorescent proteins, which allow screening of a high number of compounds in a fast and reproducible way[27-31]. However, the results from these systems should be interpreted with care because some adaptive mutations that mediate efficient replication in vitro might result in non-replicative replicons in vivo.
It has been previously reported that HCV NS3/4A protease cleaves MAVS from mitochondria to block the activation of IFN-β and nuclear factor-κB and the induction of antiviral effector genes in infected cells. In the present study, using the eYFP-MAVS/IFN-β-SEAP signaling pathway and HCV replicon model, we established a rapid and sensitive cell-based assay system to identify regulators of HCV replication. It allowed us to monitor HCV replication both qualitatively and quantitatively, using an easy, rapid and inexpensive system. HCV replication can be measured by the reporter enzyme SEAP, which enabled us to perform ongoing monitoring of anti-HCV drugs through repeated chemiluminescence. The cell lysates need not be prepared in our assay, and the SEAP protein is very stable and highly sensitive in enzymatic assays. SEAP activity in culture medium obtained from control cells transfected with eYFP-MAVS and IFN-β-SEAP was approximate 700 times greater than that obtained from replicon cells. Another reporter, eYFP-MAVS, which was localized to the mitochondrial membrane, but in the presence of HCV NS3/4A protease, it was proteolytically cleaved and shifted from the mitochondrial membrane to the cytoplasm, which can be dynamically monitored by fluorescence microscopy. Time- and dose-dependent studies have revealed that a decrease of SEAP activity and cleavage of eYFP-MAVS occur under conditions in which NS3/4A is active. Valuation of this reporter assay was verified through detecting the activity of IFN-α and CsA, which have been reported to be capable of modulating HCV replication. Although not investigated here, we believe that this reporter system could be of significant benefit in studies of HCV infection. This system will enable development or enhancement of therapeutic protocols by providing a platform to investigate dosing, scheduling, or the efficacy of combination therapies.
Advances in hepatitis C virus (HCV) replicon and infectious cell models provide systems for screening compounds against HCV. However, methods for detection of HCV protein or RNA require fixation or cell lysis, such as immunofluorescence, Western blotting and quantitative reverse transcriptase-polymerase chain reaction, and are insensitive or complex and time-consuming.
Overexpression of mitochondrial antiviral signaling protein (MAVS) strongly activates interferon (IFN)-β promoter. MAVS self-association mediates innate immune signaling that can be blocked by HCV NS3/4A cleavage. Recent studies have highlighted the mechanism by HCV escapes the innate antiviral immunity mediated by HCV NS3/4A cleavage of MAVS. In this study, the authors developed a sensitive, simple assay for screening compounds against HCV based on HCV NS3/4A cleavage of the MAVS/IFN-β signaling pathway.
Although many studies have focused on the mechanism by which HCV escapes the innate antiviral immunity mediated by HCV NS3/4A blockage of the MAVS/IFN-β signaling pathway, its application for HCV detection has seldom been studied. The assay reported here based on HCV NS3/4A blockage of the MAVS/IFN-β signaling pathway converts the theoretical research into applied science. The reporter system provides a quantitative approach for detection of HCV replication based on measurement of secreted reporters in the medium, which is more sensitive and simpler compared with the methods described in background.
The reporter system here is a simple, sensitive and quantitative assay for anti-HCV compounds and will provide another alternative efficient means for screening HCV inhibitors.
Measurement of reporter expression level regulated by IFN-β promoter is an indicator for IFN-β promoter activation. Overexpression of MAVS strongly activates IFN-β-promoter-regulated reporter expression. However, in the presence of HCV NS3/4A, IFN-β promoter activation induced by MAVS was inhibited. By contrast, compounds against HCV will recover IFN-β promoter activation induced by MAVS.
Fu et al have described concisely an HCV replicon model that is suitable to test the efficacy of anti-HCV compounds by means of checking the activity of the MAVS/IFN-β pathway. The materials and methods are clearly presented, although they could be explained in more detail. Results are demonstrative and conclusive and the English is also good, with only minor changes being necessary.
Peer reviewer: Juan-Ramón Larrubia, PhD, Gastroenterology Unit and Liver Research Unit., Guadalajara University Hospital, Donante de Sangre s/n, 19002 Guadalajara, Spain
S- Editor Tian L L- Editor Kerr C E- Editor Lin YP
| 1. | Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556-562. |
| 2. | Conry-Cantilena C, VanRaden M, Gibble J, Melpolder J, Shakil AO, Viladomiu L, Cheung L, DiBisceglie A, Hoofnagle J, Shih JW. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N Engl J Med. 1996;334:1691-1696. |
| 3. | Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558-567. |
| 4. | Husa P, Husova L. Treatment of chronic hepatitis C patients with combination of alpha-interferon and ribavirin, consensus and pegylated interferons. Bratisl Lek Listy. 2001;102:248-252. |
| 5. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. |
| 6. | De Francesco R, Carfí A. Advances in the development of new therapeutic agents targeting the NS3-4A serine protease or the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Adv Drug Deliv Rev. 2007;59:1242-1262. |
| 7. | De Francesco R, Tomei L, Altamura S, Summa V, Migliaccio G. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res. 2003;58:1-16. |
| 8. | Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453-463. |
| 9. | Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5-19. |
| 10. | Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017-4026. |
| 11. | Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299-1311. |
| 12. | Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103:6001-6006. |
| 13. | Gale M Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939-945. |
| 14. | Foy E, Li K, Wang C, Sumpter R Jr, Ikeda M, Lemon SM, Gale M Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145-1148. |
| 15. | Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992-2997. |
| 16. | Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795-1803. |
| 17. | Ciesek S, Steinmann E, Wedemeyer H, Manns MP, Neyts J, Tautz N, Madan V, Bartenschlager R, von Hahn T, Pietschmann T. Cyclosporine A inhibits hepatitis C virus nonstructural protein 2 through cyclophilin A. Hepatology. 2009;50:1638-1645. |
| 18. | Watashi K, Shimotohno K. Chemical genetics approach to hepatitis C virus replication: cyclophilin as a target for anti-hepatitis C virus strategy. Rev Med Virol. 2007;17:245-252. |
| 19. | Nakagawa M, Sakamoto N, Enomoto N, Tanabe Y, Kanazawa N, Koyama T, Kurosaki M, Maekawa S, Yamashiro T, Chen CH. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem Biophys Res Commun. 2004;313:42-47. |
| 20. | Bartenschlager R, Kaul A, Sparacio S. Replication of the hepatitis C virus in cell culture. Antiviral Res. 2003;60:91-102. |
| 21. | Boonstra A, van der Laan LJ, Vanwolleghem T, Janssen HL. Experimental models for hepatitis C viral infection. Hepatology. 2009;50:1646-1655. |
| 22. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. |
| 23. | Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci USA. 2006;103:3805-3809. |
| 24. | Liu C. Hepatitis C virus: virology and experimental systems. Clin Liver Dis. 2006;10:773-791. |
| 25. | Horscroft N, Lai VC, Cheney W, Yao N, Wu JZ, Hong Z, Zhong W. Replicon cell culture system as a valuable tool in antiviral drug discovery against hepatitis C virus. Antivir. Chem Chemother. 2005;16:1-12. |
| 26. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. |
| 27. | Zhang Y, Weady P, Duggal R, Hao W. Novel chimeric genotype 1b/2a hepatitis C virus suitable for high-throughput screening. Antimicrob Agents Chemother. 2008;52:666-674. |
| 28. | Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308-5320. |
| 29. | Hao W, Herlihy KJ, Zhang NJ, Fuhrman SA, Doan C, Patick AK, Duggal R. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob Agents Chemother. 2007;51:95-102. |
| 30. | Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol. 2004;78:7400-7409. |
| 31. | Jones DM, Gretton SN, McLauchlan J, Targett-Adams P. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J Gen Virol. 2007;88:470-475. |