Published online Jul 7, 2010. doi: 10.3748/wjg.v16.i25.3153
Revised: April 21, 2010
Accepted: April 28, 2010
Published online: July 7, 2010
AIM: To understand CD133 promoter hypermethylation and expression in 32 colorectal cancer cell lines.
METHODS: Nucleic acid was isolated from 32 colorectal cancer cell lines and CD133 expression levels were measured by reverse transcription-polymerase chain reaction (RT-PCR) and real-time PCR. Promoter methylation status of the CD133 gene was analyzed with a methylation-specific PCR after sodium-bisulfite modification and by clonal sequencing analysis. The correlation between expression and promoter methylation of CD133 gene was confirmed with treatment of 5-aza-2’-deoxycytidine.
RESULTS: We measured CD133 expression levels in 32 colorectal cancer cell lines. RT-PCR analysis showed undetectable or low levels of CD133 expression in 34.4% of cell lines. To verify the relation between CD133 expression and methylation status of the CD133 gene promoter in colorectal carcinogenesis, CD133 gene promoter hypermethylation was analyzed in 32 cancer cell lines. Promoter hypermethylation was detected in 13 (40.6%) of the cell lines using methylation specific-PCR and confirmed by bisulfite sequencing analysis. Treatment of 11 of the cell lines with the demethylation agent 5-aza-2’-deoxycytidine recovered CD133 expression in most of them.
CONCLUSION: Transcriptional repression of CD133 is caused by promoter hypermethylation of the CD133 CpG islands in some of colorectal cancer cell lines. The study may contribute to the understanding of the role of CD133 inactivation in the progression of colorectal cancers.
-
Citation: Jeon YK, Kim SH, Choi SH, Kim KH, Yoo BC, Ku JL, Park JG. Promoter hypermethylation and loss of
CD133 gene expression in colorectal cancers. World J Gastroenterol 2010; 16(25): 3153-3160 - URL: https://www.wjgnet.com/1007-9327/full/v16/i25/3153.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i25.3153
The cell surface antigen CD133 is a glycoprotein of 858-865 amino acids with a total molecular weight of 97-120 kDa and five transmembrane domains. Originally identified in neuroepithelial stem cells, CD133 has been detected in hematopoietic stem cells, endothelial progenitor cells, glioblastomas, neuronal and glial stem cells, and some other cell types[1,2]. As it has also been identified in cancer-initiating cells in several solid malignancies, it is regarded as one of the cancer stem cell (CSC) markers in colorectal carcinoma[3,4]. CD133 was the first identified member of a pentaspan membrane protein family in both humans and mice[1]. The glycoprotein specifically localizes in microvilli and protruding plasma membrane[5].
The CSC theory is a newly emerged concept of cancer initiation and development. According to this theory, only a small population of cells is clonogenic and contains tumor initiating potency, whereas the majority of the tumor cells have undergone differentiation and lost this potency. In colorectal cancer, these cells have been reported to express CD133 and a CD133-positive population of colon cancer cells was recently demonstrated to be highly enriched in tumor-initiating colon CSCs that have the ability to self-renew and to recapitulate the bulk tumor population[3,6-9].
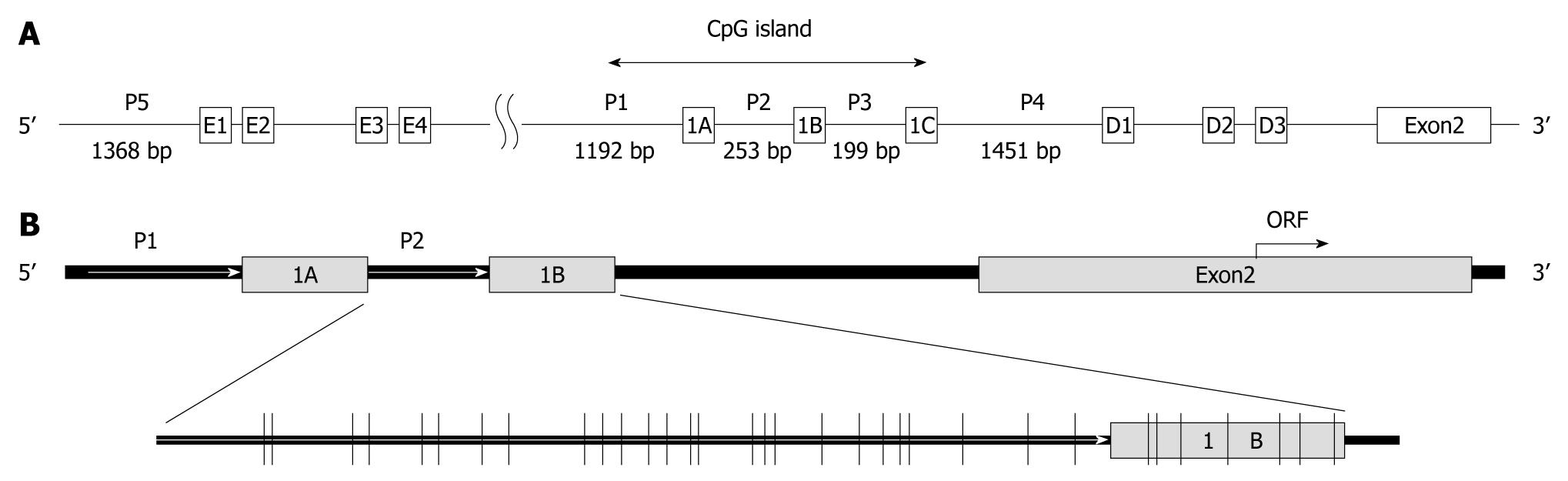
The CD133 gene transcriptional regulation is rather complicated and poorly understood. A possible involvement of five alternative TATA-less promoters has been suggested to explain the pattern of transcripts differing in the lengths and sequences of 5′ untranslated regions (UTRs). Two of these promoters, P1 and P2 (Figure 1A), are active in in vitro tests with a reporter gene. A common transcriptional initiation site was assigned to exons 1A and 1B (Figure 1A), and an mRNA transcribed from exon 1A or 1B was found to be the major transcript, with the choice between the transcription start sites depending on tissues. In particular, colon-expressed transcripts contain both exons 1A and 1B[10-12].
Changes in DNA methylation patterns are an important hallmark of tumor development and progression. Methylation of the C5 position of cytosine residues in DNA is one of the most fundamental epigenetic characteristics. This methylation is performed by DNA methyl-transferases (DNMTs), which have been implicated in many processes including transcriptional regulation, genomic stability, X chromosome inactivation and silencing of parasitic DNA transposable elements[13]. The importance of DNA methylation is highlighted by the finding that many human diseases result from its abnormal control[14]. Moreover, the aberrant methylation of CpG islands is characteristic of many human cancers and is detected during early carcinogenesis[15]. Hypermethylation of promoter CpG islands is the signature of transcriptional silencing of tumor suppressor genes in various human cancers, and this is as effective as inactivation by gene mutation or deletion[16,17].
To examine whether CD133 expression is related to promoter methylation of the gene, we assessed the expression of the CD133 gene in 32 colorectal cancer cell lines and determined the methylation status of the CD133 promoter in each cell line.
A total of 32 colorectal cancer cell lines were obtained from the Korean Cell Line Bank (KCLB; Seoul, Korea). Sixteen SNU-colorectal cancer cell lines were established as previously reported by our laboratory[18]. All the cell lines were maintained in RPMI1640 medium except for two cell lines; Caco-2 and WiDr were maintained in Minimum Essential Medium and Dulbecco’s modified Eagle’s medium, respectively. The media were supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 0.1 mg/mL streptomycin. Cultures were maintained in humidified incubators at 37°C in a 5% CO2 and 95% air atmosphere. All cell lines were absent of mycoplasma (e-myco mycoplasma PCR detection kit, Intron Biotechnology, Gyeonggi, Korea) and bacteria contamination and genetic heterogeneity by DNA fingerprinting analysis (AmpFlSTR Identifiler PCR amplification kit, Applied Biosystems, Foster City, CA, USA).
Genomic DNA and total RNA were isolated from washed cell pellets. Genomic DNA was extracted using a G-DEX™ kit (Intron Biotechnology) according to the manufacturer’s instructions. Total RNA was isolated according to the manufacturer’s instructions using easy-BLUE™ kits (Intron Biotechnology). For cDNA synthesis, 2 μg of total RNA was reversely transcribed using a random primer, dNTP, and 1 μL (200 units) of Superscript™ II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) in a final volume of 20 μL for 80 min at 42°C after a 15-min denaturation at 80°C. Eighty microliters of distilled water was then added to the reverse-transcription reaction.
For CD133 expression analysis, the cDNA was amplified in 25 μL of a PCR reaction mixed with 1 μL of the reverse-transcription reaction, primers and 1 unit of Taq DNA polymerase. Reverse transcription-polymerase chain reaction (RT-PCR) was carried out using RT specific primers (located +343 to +679 from the translation start site in the mRNA sequence); CD133 RT sense (5'-CTGGGGCTGCTGT TTATTATTCTG-3'), and CD133 RT antisense (5'-ACGCCTTGTCCTTGGTAGTGTTG-3'). PCR conditions consisted of 5 min at 94°C for initial denaturation, followed by 35 cycles of 94°C (30 s), 55°C (30 s), and 72°C (60 s) and a final elongation of 7 min at 72°C. PCR amplification was performed in a programmable thermal cycler (PCR System 9700; Applied Biosystems; Foster City, CA, USA). Primers for β-actin were used to confirm RNA integrity. Both CD133 and β-actin RT-PCR reactions used the same cDNA synthesis. The amplified DNA fragments were fractionated in 2% agarose gel and stained with ethidium bromide.
Real-time PCR was performed in 386 well PCR plates containing the 2X FastStart Universal SYBR Green Master (ROX) mix (Roche, Basel, Switzerland), 10 ng of cDNA template, 300 nmol/L of CD133 RT sense primer and 600 nmol/L of CD133 RT antisense primer in a final volume of 10 μL. Each primer/cDNA set was set up in triplicate. Real-time PCR reactions in a 7900HT Fast Real-Time PCR System (Applied Biosystems) were initiated by heating to 50°C for 2 min and then to 95°C for 10 min, followed by 40 cycles of 95°C (15 s), and 60°C (60 s). The relative quantification of gene expression was performed using the standard curve method. The standard curve for CD133 expression level was constructed using serial dilutions of SNU-407 cDNA, which in preliminary experiments displayed strong RT-PCR expression of the CD133 gene. Samples normalized to β-actin served as an internal control.
For methylation analysis, 2 μg of genomic DNA obtained from colorectal cancer cell lines was modified using EZ DNA Methylation™ Kit (Zymo Research, Orange, CA, USA). The primers specific for bisulfite modified DNA were designed using MethPrimer software (http://www.urogene.org/methprimer/index1.html). The used primers (located -8061 to -7782 from transcription start site) were as follows; CD133 M primer sense (5'-TTCGGGATAGAGGAAGTCGTAA-3'), CD133 M primer antisense (5'-CTCCCGCCCTAATCACCGCT-3'), CD133 U primer sense (5'-TTTGGGATAGAGGAAGTTGTAA-3') and CD133 U primer antisense (5'-CTCCCACCCTAATCACCACT-3'). The PCR conditions were as follows : 94°C for 5 min, and then 43 cycles of 94°C for 30 s, 60°C(methylation specific PCR, MSP) or 62°C (unmethylation specific PCR, USP) for 30 s, and 72°C for 60 s, and finally 72°C for 7 min. The amplified DNA fragments were fractionated in a 2% agarose gel and stained with ethidium bromide.
Sequencing primers recognizing both methylated and unmethylated sites (MU), CD133 MU sense (5'-TATTTGGTTATGTTTTTAGTTTTTT-3') and CD133 MU antisense (5'-CCTAATCAACAAATACCTCTCTC-3') primers also were designed using the MethPrimer software. These primers were located approximately -8103 to -7708 from transcription start site. The PCR conditions used were 5 min at 94°C for initial denaturation, 40 amplification cycles of 94°C (30 s), 59°C (30 s) and 72°C (60 s) and a final elongation of 7 min at 72°C. The PCR products obtained with bisulfite sequencing primers were inserted into the pGEM-T Easy vector (Promega, Madison, WI, USA) for cloning. Sequences of five individual colonies for each analyzed cell line were determined using universal pUC/M13 primers and each sequence was analyzed using a Taq dideoxy terminator cycle sequencing kit on an ABI 3730 DNA sequencer (Applied Biosystems).
For treatment with 5-aza-2’-deoxycytidine, 2 × 105 cells were seeded in two 75 cm2 culture flasks on day 0. The cells were untreated or treated with 5 μmol/L of 5-aza-2’-deoxycytidine (Sigma-Aldrich, St. Louis, MO, USA) for 24 h on day 2. The culture was re-dosed every 48 h (days 4 and 6) and medium was changed 24 h after adding 5-aza-2’-deoxycytidine. The cells were harvested on day 8 for RNA isolation. The RNA was used for cDNA synthesis and analysis of the CD133 expression as described above.
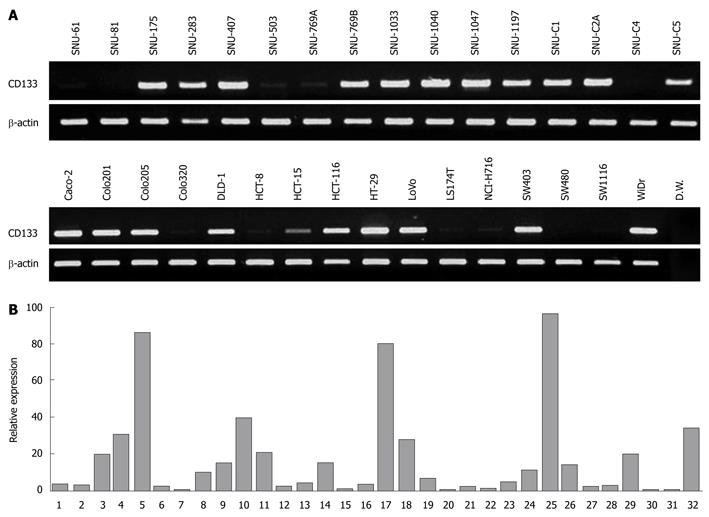
We analyzed expression of CD133 in 32 colorectal cancer cell lines by both RT-PCR and quantitative real-time PCR. In RT-PCR analysis, CD133 expression was observed in 21 of the 32 cell lines (65.6%) (Figure 2A). On the other hand, in 11 cell lines (34.4%), CD133 expression was either undetectable (SNU-81 and SW480) or low (SNU-61, SNU-503, SNU-769A, SNU-C4, Colo320, HCT-8, LS174T, NCI-H716 and SW1116). To verity the RT-PCR results, we performed real-time PCR and quantified CD133 expression against β-actin expression. All 11 cell lines showed low relative expression (< 3.5%) (Figure 2B). Real-time PCR also demonstrated that 16 of 20 cell lines having strong expression of the CD133 gene displayed high relative expression (> 6%); the four exceptions were SNU-1197, SNU-C1, SNU-C5 and DLD-1.
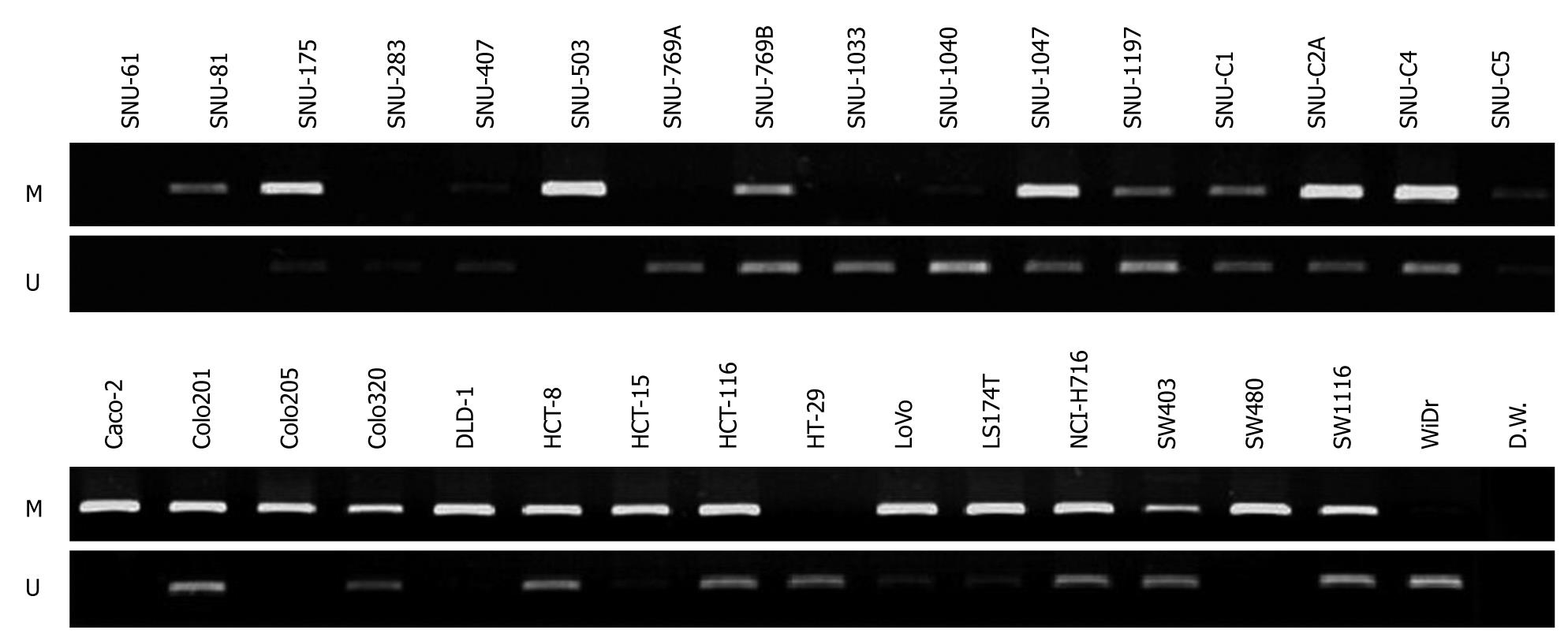
To assess if CD133 gene expression was influenced by methylation of its promoter, we checked whether promoter CpG islands of the gene were methylated or unmethylated in the 32 colorectal cancer cell lines by performing methylation specific-PCR (MS-PCR) with designed methylation and unmethylation primers (Figure 3). Methylated DNAs were amplified in 26 cell lines (SNU-81, SNU-175, SNU-407, SNU-503, SNU-769B, SNU-1040, SNU-1047, SNU-1197, SNU-C1, SNU-C2A, SNU-C4, SNU-C5, Caco-2, Colo201, Colo205, Colo320, DLD-1, HCT-8, HCT-15, HCT116, LoVo, LS174T, NCI-H716, SW403, SW480 and SW1116) and unmethylated DNAs were amplified in 24 cell lines (SNU-175, SNU-283, SNU-407, SNU-769A, SNU-769B, SNU-1033, SNU-1040, SNU-1047, SNU-1197, SNU-C1, SNU-C2A, SNU-C4, SNU-C5, Colo201, Colo320, HCT-8, HCT-15, HCT116, HT-29, LoVo, LS174T, NCI-H716, SW403, SW1116 and WiDr). Only methylated DNA bands were evident in SNU-81 and SW480, with undetectable CD133 expression, and in the SNU-503 cell line, with weak CD133 gene expression. In contrast, only unmethylated DNA bands were evident in SNU-283, SNU-1033, HT-29, and WiDr, in which CD133 gene expression was strong. Both methylated and unmethylated DNA bands were detected in 13 cell lines (SNU-175, SNU-407, SNU-769B, SNU-1040, SNU-1047, SNU-1197, SNU-C1, SNU-C2A, SNU-C5, Colo201, HCT116, LoVo and SW403), which also showed strong expression of the CD133 gene. Three cell lines (Caco-2, Colo205 and DLD-1) contained only methylated DNA bands, even though these cell lines expressed CD133. Methylated or unmethylated DNA bands were not evident in SNU-61 cell line.
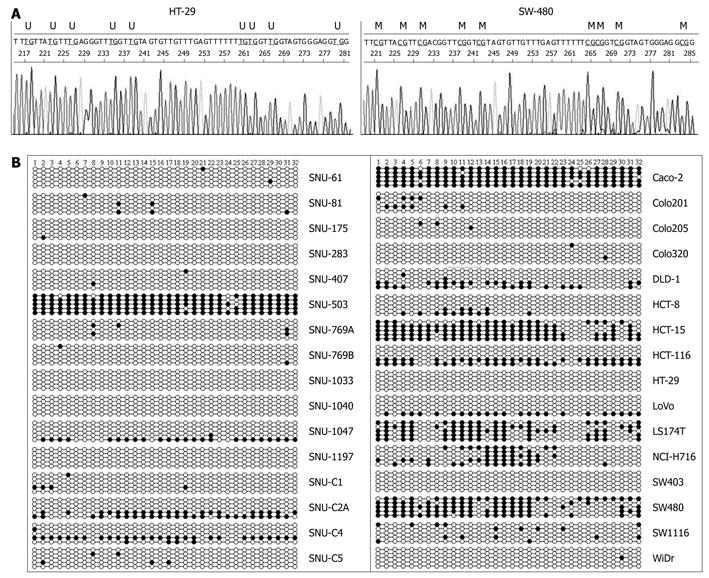
A study reported that methylation of promoter P1 and exon A1 did not correlate with the CD133 gene transcription level because promoter P1 and exon 1A were hypermethylated with a high (90%-94%) content of methylated CpG dinucleotides in all cell lines examined[11]. Appropriately, we investigated the methylation status of 32 CpG sites in promoter P2 and exon 1B (Figure 1B) relative to the transcription initiation site of CD133 gene by clonal bisulfite sequencing analysis. Representative sequence diagrams of two cell lines are shown in Figure 4A. The methylation status of promoter CpG dinucleotides (Figure 4B) was correlated with CD133 expression (Table 1). The promoter CpG dinucleotides of CD133 of 13 cell lines (SNU-175, SNU-283, SNU-407, SNU-769B, SNU-1033, SNU-1040, SNU-1197, SNU-C1, SNU-C5, Colo201, Colo205, HT-29, SW403 and WiDr) were mostly unmethylated. Hypermethylation of CpG islands was 64% in the SW480 cell line (undetectable CD133 gene expression), 64%-95% in SNU-503, HCT-15 and LS174T cell lines (low expression of CD133 gene) and 9%-24% in SNU-C4, HCT-8, NCI-H716 and SW1116 cell lines (also low expression of CD133 gene). The distribution of methylated CpG dinucleotides in different clones was not uniform. For example, in SNU-C2A cells the methylation level of CpG in promoter was markedly increased through hypermethylation in two clones, whereas the CpG dinucleotides in other clones were fully unmethylated (Figure 4B). Similar variations were observed in SNU-1047, DLD-1, HCT116, and LoVo cell lines. The variance was suggestive of the origin of different clones from different alleles of the gene. However, further studies are needed to confirm this possibility.
| Cell lines (32 in total) | M | U | % methylation | % expression |
| SNU-61 | - | - | 1 | 3.1 |
| SNU-81 | + | + | 4 | 2.8 |
| SNU-175 | ++ | + | 1 | 19.9 |
| SNU-283 | - | + | 0 | 30.7 |
| SNU-407 | + | + | 1 | 86.2 |
| SNU-503 | ++ | - | 95 | 2.4 |
| SNU-769A | - | + | 3 | 0.7 |
| SNU-769B | + | ++ | 1 | 10.2 |
| SNU-1033 | - | + | 0 | 14.7 |
| SNU-1040 | + | ++ | 0 | 40.2 |
| SNU-1047 | ++ | + | 16 | 20.3 |
| SNU-1197 | + | ++ | 0 | 2.4 |
| SNU-C1 | + | + | 3 | 4.0 |
| SNU-C2A | ++ | + | 29 | 14.7 |
| SNU-C4 | ++ | ++ | 21 | 0.5 |
| SNU-C5 | + | + | 3 | 3.7 |
| Caco-2 | ++ | - | 95 | 79.6 |
| Colo201 | ++ | ++ | 6 | 27.4 |
| Colo205 | ++ | - | 2 | 6.8 |
| Colo320 | ++ | + | 1 | 0.7 |
| DLD-1 | ++ | - | 21 | 2.4 |
| HCT-8 | ++ | ++ | 9 | 1.4 |
| HCT-15 | ++ | + | 73 | 4.7 |
| HCT-116 | ++ | ++ | 31 | 11.2 |
| HT-29 | - | ++ | 0 | 96.0 |
| LoVo | ++ | + | 14 | 13.9 |
| LS174T | ++ | + | 53 | 2.4 |
| NCI-H716 | ++ | ++ | 24 | 2.6 |
| SW403 | + | ++ | 0 | 19.7 |
| SW480 | ++ | - | 64 | 0.6 |
| SW1116 | ++ | ++ | 10 | 0.7 |
| WiDr | + | ++ | 0 | 33.3 |
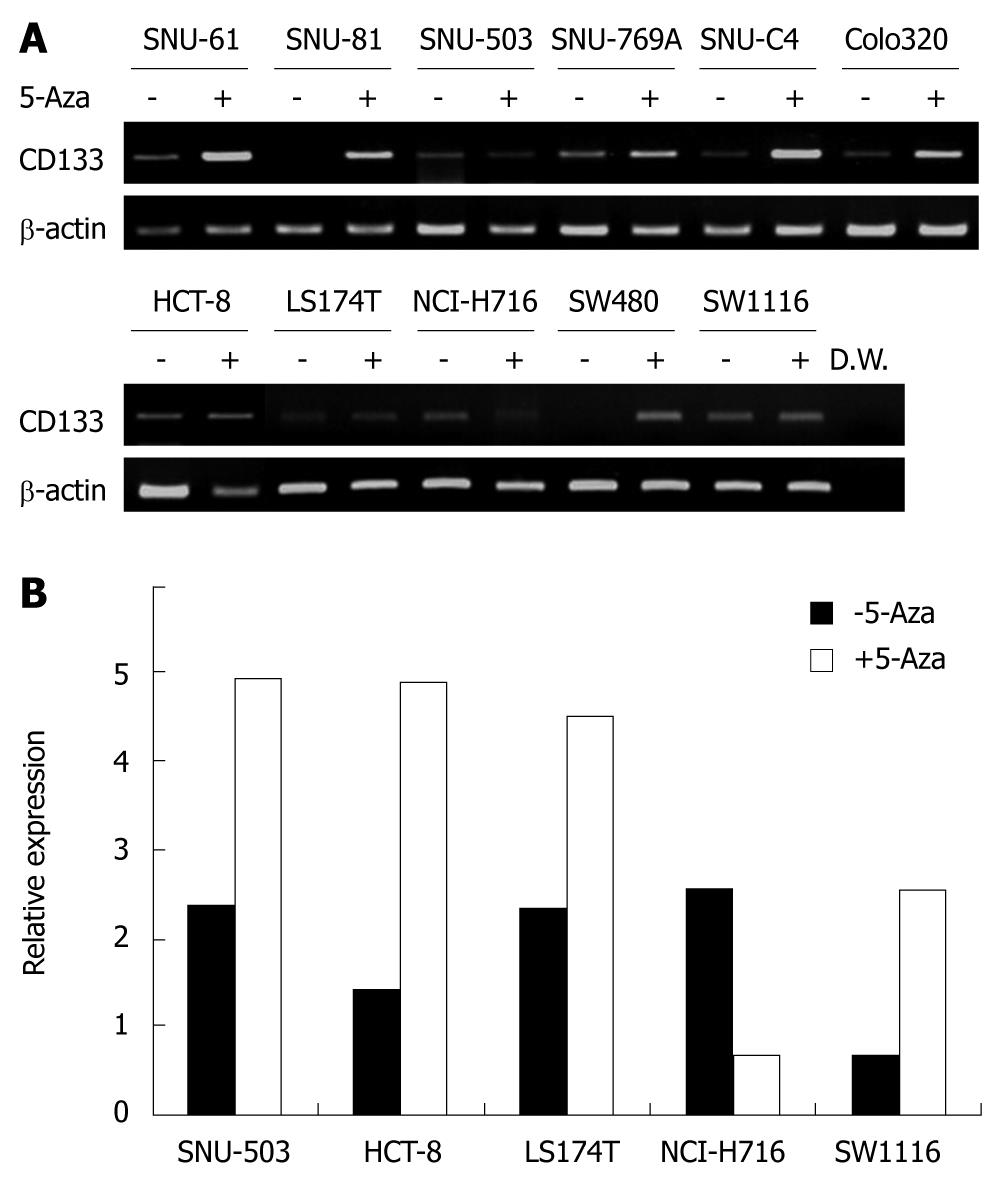
To confirm whether the differential CD133 expression was related to DNA methylation, 11 cell lines with undetectable or low expression of CD133 were treated with 5-aza-2’-deoxycytidine. The treatment recovered CD133 expression in four of the weakly CD133 expressing cell lines (SNU-61, SNU-769A, SNU-C4 and Colo320) and two cell lines without detectable CD133 expression (Figure 5A). However, 5-aza-2’-deoxycytidine treatment did not significantly influence CD133 gene expression in SNU-503, HCT-8, LS174T, NCI-H716 and SW1116 cell lines. Examination of CD133 expression by real- time PCR revealed that CD133 expression was increased after 5-aza-2’-deoxycytidine treatment in SNU-503, HCT-8, LS174T and SW1116 cell lines (Figure 5B), and decreased in the NCI-H716 cell line.
It has been postulated that CSCs are able to maintain tumor bulk due to the abilities of self-renewal and differentiation into cells with low potential. CD133 is one of CSC markers in colorectal carcinoma and CD133-positive cells in colon cancer exhibit a high tumorigenic ability in vivo. In a previous study, the CD133-positive cells separated from tumors were able to form tumor bulk in the immuno-compromised mouse models with less numbers than the CD133-negative cells and these cells showed a long-term tumorigenic potential[4]. Furthermore, CD133-positive cells in colon cancer cell lines showed higher levels of proliferation, colony formation and invasive ability in vitro than CD133-negative cells[19].
Presently, CD133 was down-regulated in 11 of the 32 colorectal cancer cell lines. Since abnormal DNA methylation of CD133 gene is related to CD133 expression in colorectal tumors[20], we hypothesize that CD133 expression is regulated by hypermethylation of the promoter region in this gene, and analyzed promoter methylation status of the CD133 gene with a methylation-specific PCR after sodium-bisulfite modification and by clonal sequencing analysis. A methylation analysis of promoters P1 and P2 in cell lines demonstrated tissue specificity of the promoter P2 methylation and practically no specificity in the methylation of the P1 promoter[11]. Therefore, we designed the primers with promoter P2 sequences for MS-PCR and bisulfite sequencing analysis and sequencing primers containing 32 of CpG dinucleotides (Figure 1B).
Methylation of the CD133 gene promoter was observed in 13 cell lines (SNU-503, SNU-1047, SNU-C2A, SNU-C4, Caco-2, DLD-1, HCT-15, HCT116, LoVo, LS174T, NCI-H716, SW403 and SW1116). These cells exhibited significant methylation bands in MS-PCR analysis, and bisulfite sequencing analysis revealed that > 10% of the total CpG islands were hypermethylated. DNA methylation in the promoter region of a gene is associated with a loss of gene expression and plays an important role in gene silencing. The inactivation of tumor-suppressor genes by aberrant methylation in the promoter region is well-recognized in carcinogenesis[20].
However, loss of CD133 expression in early colorectal cancer is different from expression loss of tumor suppressor genes. Acquisition of CD133 promoter methylation of cells without CD133 expression resulted from CD133 positive cell division. The inverse correlation between CD133 transcription and methylation provides a mechanistic explanation for the loss of cell surface CD133 expression in differentiated cells. This is consistent with the notion that cell differentiation is accompanied by epigenetic changes that are responsible for guiding the future phenotypic profile of the progeny[21]. This phenomenon is not only unique to normal stem cells but also presents aberrantly in CSCs, which may initiate carcinogenesis[22,23]. In advanced colorectal carcinoma, the CD133 gene was more frequently demethylated[24]. The carcinomas with demethylation of CD133 gene showed a bigger maximal tumor size and a trend toward the development of a lymph node metastasis.
In our results of bisulfite sequencing analysis, there are differences of methylation status among the colonies of the same cell line. The variance was suggestive of the origin of different clones from different alleles of the gene. Definitely, heterogeneity of DNA methylation for several genes has been observed in total cell populations from cultured and primary cancers. The present observations for CD133 promoter methylation are unique in showing striking heterogeneity between isolated cell populations in single-tumor culture lines. This seems to be a more uniform heterogeneity involving cells of the tumor and manifesting as quantitative differences between alleles of a given gene. These quantitative differences of abnormal promoter DNA methylation can be quantitatively altered by changes in environmental surrounding for cultured tumor cells[25].
CD133 has been re-expressed by demethylation with 5-aza-2’-deoxycytidine in some cell lines. This agent reactivates gene expression when gene expression is reduced by methylation of CpG islands. Our results confirm that inactivation of CD133 expression is related to epigenetic modification, which, in colorectal cancer cell lines, is promoter methylation. The function of CD133 is currently unknown, but it was reported that CD133 expression is repressed by DNA methylation in CD133-negative progeny of CD133-positive cells[26], supporting a role for CD133 in CD133-positive cells. It has been found that the expression pattern of several genes was changed in neurosphere cells by treatment of chromatin-modifying agents, 5-aza-2’-deoxycytidine and trichostatin A, and these cells induced hematopoietic activity in vivo[27]. Therefore, re-expression of CD133 by treatment of demethylation agent is expected to discover the functions of CD133 gene in cancer.
The failure to detect methylated or unmethylated DNA bands in MS-PCR SNU-61 cells warrants comment. We believe that there are some problems with the quality of the modified DNA. This remains to be confirmed. The strong expression of CD133 by Caco-2 cells, in which promoter hypermethylation was detected, supports the suggestion that regulation of CD133 expression might be caused by another mechanism.
In conclusion, we observed hypermethylation in the promoter region of the CD133 gene in 13 of 32 colorectal cancer cell lines. We confirmed the methylation status by MS-PCR, bisulfite sequencing analysis and treatment of 5-aza-2’-deoxycytidine. The expression status of the CD133, one of CSC markers, was correlated with methylation status of CpG islands in the CD133 promoter. These results may contribute to the understanding of the role of CD133 inactivation in the pathogenesis of colorectal cancers.
The caner stem cell theory is a newly emerged concept of cancer initiation and development. These cells have the ability to self-renew and to recapitulate the bulk tumor population. In colorectal cancer, a CD133-positive population of colon cancer cells was recently demonstrated to be highly enriched in tumor-initiating colon cancer stem cells.
It is reported that tumor initiating cells in colorectal cancer cells express CD133, the cell surface glycoprotein. However, the CD133 gene transcriptional regulation is rather complicated and poorly understood. This study demonstrates that CD133 expression could be regulated by methylation status of promoter of the gene.
Recently, a CD133 has become a matter of common interest in the research of colorectal cancer stem cells. This study suggests that transcriptional repression of CD133 is caused by promoter hypermethylation of the CD133 in some of colorectal cancer cell lines. Moreover, each colorectal cancer cell line represented different methylation status of promoter and each colony of cell lines exhibited different levels of methylation.
Cancer stem cells lost their potential during differentiation by loss of expression of stem cell-specific expressed genes. Therefore, this study may contribute to the understanding of the role of CD133 inactivation in the progression of colorectal cancers.
Promoter methylation is one of the most essential epigenetic characteristics. The importance of DNA methylation is highlighted by the finding that many human diseases result from its abnormal control. Moreover, the aberrant methylation of CpG islands is characteristic of many human cancers and is detected during early carcinogenesis.
This manuscript investigates the promoter hypermethylation and expression status of CD133, which is one of several putative cancer stem cell markers in colon cancer. The overall findings indicate that there is a correlation between this epigenetic change in some cancer cell lines, and that demethylation is capable of restoring CD133 expression in some cell lines. Furthermore, there is a surprising finding that different colonies of a particular cell line exhibited different levels of methylation, indicating as the authors point out, that other influences (e.g. tumor environment) may be involved.
Peer reviewer: Baljinder Singh Salh, MRCP, LMCC, FRCP(C), Associate Professor, University of British Columbia, 5th Floor, 2775 Laurel Street, Vancouver, V5Z1M9, Canada
S- Editor Wang YR L- Editor Ma JY E- Editor Ma WH
| 1. | Corbeil D, Röper K, Hellwig A, Tavian M, Miraglia S, Watt SM, Simmons PJ, Peault B, Buck DW, Huttner WB. The human AC133 hematopoietic stem cell antigen is also expressed in epithelial cells and targeted to plasma membrane protrusions. J Biol Chem. 2000;275:5512-5520. |
| 2. | Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 2005;37:715-719. |
| 3. | O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. |
| 4. | Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-115. |
| 5. | Tabu K, Sasai K, Kimura T, Wang L, Aoyanagi E, Kohsaka S, Tanino M, Nishihara H, Tanaka S. Promoter hypomethylation regulates CD133 expression in human gliomas. Cell Res. 2008;18:1037-1046. |
| 6. | Burkert J, Wright NA, Alison MR. Stem cells and cancer: an intimate relationship. J Pathol. 2006;209:287-297. |
| 7. | Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267-284. |
| 8. | Vermeulen L, Sprick MR, Kemper K, Stassi G, Medema JP. Cancer stem cells--old concepts, new insights. Cell Death Differ. 2008;15:947-958. |
| 9. | Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427-13432. |
| 10. | Katoh Y, Katoh M. Comparative genomics on PROM1 gene encoding stem cell marker CD133. Int J Mol Med. 2007;19:967-970. |
| 11. | Pleshkan VV, Vinogradova TV, Sverdlov ED. Methylation of the prominin 1 TATA-less main promoters and tissue specificity of their transcript content. Biochim Biophys Acta. 2008;1779:599-605. |
| 12. | Shmelkov SV, Jun L, St Clair R, McGarrigle D, Derderian CA, Usenko JK, Costa C, Zhang F, Guo X, Rafii S. Alternative promoters regulate transcription of the gene that encodes stem cell surface protein AC133. Blood. 2004;103:2055-2061. |
| 13. | Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA. 2005;102:3336-3341. |
| 14. | Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597-610. |
| 15. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. |
| 16. | Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 Suppl 1:S4-S11. |
| 17. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. |
| 18. | Park JG, Oie HK, Sugarbaker PH, Henslee JG, Chen TR, Johnson BE, Gazdar A. Characteristics of cell lines established from human colorectal carcinoma. Cancer Res. 1987;47:6710-6718. |
| 19. | Ieta K, Tanaka F, Haraguchi N, Kita Y, Sakashita H, Mimori K, Matsumoto T, Inoue H, Kuwano H, Mori M. Biological and genetic characteristics of tumor-initiating cells in colon cancer. Ann Surg Oncol. 2008;15:638-648. |
| 20. | Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, Yoshikawa T, Yamanaka S, Endo C, Sakurada A, Sato M. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059-3064. |
| 21. | Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2-9. |
| 22. | Balch C, Nephew KP, Huang TH, Bapat SA. Epigenetic "bivalently marked" process of cancer stem cell-driven tumorigenesis. Bioessays. 2007;29:842-845. |
| 23. | Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237-242. |
| 24. | Hibi K, Sakata M, Kitamura YH, Sakuraba K, Shirahata A, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G. Demethylation of the CD133 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009;29:2235-2237. |
| 25. | Yi JM, Tsai HC, Glöckner SC, Lin S, Ohm JE, Easwaran H, James CD, Costello JF, Riggins G, Eberhart CG. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 2008;68:8094-8103. |
| 26. | Baba T, Convery PA, Matsumura N, Whitaker RS, Kondoh E, Perry T, Huang Z, Bentley RC, Mori S, Fujii S. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28:209-218. |
| 27. | Ruau D, Ensenat-Waser R, Dinger TC, Vallabhapurapu DS, Rolletschek A, Hacker C, Hieronymus T, Wobus AM, Müller AM, Zenke M. Pluripotency associated genes are reactivated by chromatin-modifying agents in neurosphere cells. Stem Cells. 2008;26:920-926. |