Published online Jun 14, 2010. doi: 10.3748/wjg.v16.i22.2754
Revised: December 2, 2009
Accepted: December 9, 2009
Published online: June 14, 2010
AIM: To identify differentially expressed hydrophobic proteins in colorectal cancer.
METHODS: Eighteen pairs of colorectal cancerous tissues in addition to tissues from normal mucosa were analysed. Hydrophobic proteins were extracted from the tissues, separated using 2-D gel electrophoresis and analysed using Liquid Chromatography Tandem Mass Spectrometry (LC/MS/MS). Statistical analysis of the proteins was carried out in order to determine the significance of each protein to colorectal cancer (CRC) and also their relation to CRC stages, grades and patients’ gender.
RESULTS: Thirteen differentially expressed proteins which were expressed abundantly in either cancerous or normal tissues were identified. A number of these proteins were found to relate strongly with a particular stage or grade of CRC. In addition, the association of these proteins with patient gender also appeared to be significant.
CONCLUSION: Stomatin-like protein 2 was found to be a promising biomarker for CRC, especially in female patients. The differentially expressed proteins identified were associated with CRC and may act as drug target candidates.
- Citation: Yeoh LC, Loh CK, Gooi BH, Singh M, Gam LH. Hydrophobic protein in colorectal cancer in relation to tumor stages and grades. World J Gastroenterol 2010; 16(22): 2754-2763
- URL: https://www.wjgnet.com/1007-9327/full/v16/i22/2754.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i22.2754
Cancer of the colon and rectum is termed colorectal cancer (CRC). CRC is ranked the third most common cancer worldwide. Statistical data from the World Cancer Report revealed that there are more than 940 000 cases of CRC each year with an annual mortality of around 500 000[1]. In Malaysia, CRC is the most common type of cancer amongst men and the 3rd most common cancer amongst women after breast and cervical cancer[2].
Proteomics in cancer is used to study the protein expression pattern of normal and cancerous tissues in order to elucidate the molecular mechanism associated with disease development and progression[3]. Membrane proteins which make up approximately 30% of the human proteome are important components of cells[4]. Membrane-associated proteins are involved in various fundamental biological processes in cells including signal transduction, immune regulation and transportation[5]. Nevertheless, the hydrophobic nature of membrane proteins make the extraction of these proteins challenging as they are difficult to extract and analyse[6]. Solubilisation of membrane proteins requires the use of stringent reagents and the addition of urea and thiourea[7,8]. Moreover, these proteins are prone to precipitation at their isoelectric points which may lead to protein loss during 2D-Polyacrylamide Gel electrophoresis (2D-PAGE) separation[9,10]. Thus, the proteome of membrane proteins is normally under-represented. Many investigations have been carried out to improve extraction, enrichment and separation of membrane and membrane-associated proteins[11,12].
In this study, a mixture of reagents was added to the extraction buffer in order to increase the solubility of membrane proteins. Although it is impossible to extract all the membrane proteins from tissues, we obtained a consistent pattern of protein maps for cancerous and normal tissues, respectively. This enabled us to identify unique or differentially expressed membrane proteins in CRC tissues which may be useful for the diagnosis or treatment of CRC. The significance of these proteins in relation to cancer stages, grades and patient gender was also evaluated.
Surgically removed colorectal cancerous tissues and their respective normal mucosa were obtained from 18 patients who received treatment at Penang General Hospital, Malaysia. Written informed consent was obtained from all patients before surgery. The tissue specimens were kept in -80°C until analysis. The tissues were grouped according to cancer stage, grade and patient gender (Table 1). The tumor type obtained was adenocarcinoma and normal mucosa was obtained from a site at least 10 cm away from the tumor. None of the patients received preoperative neo-adjuvant chemotherapy or radiotherapy. All the patients were > 40 years old at the time of surgery. The tissues were pathologically confirmed by the hospital’s pathologist. Frozen sections of tissue were taken from cancerous tissues in the anterior and deep region to ensure adequacy of tumor and only cancerous tissue that contained > 90% malignant cells was used in this study.
| No. | Age (yr) | Sex | Duke’s system | Localization | Degree of differentiation |
| 1 | 57 | F | C | Right colon | Well-differentiated |
| 2 | 64 | M | C | Caecum | Moderately-differentiated |
| 3 | 45 | M | B | Sigmoid colon | Well-differentiated |
| 4 | 91 | F | C | Sigmoid colon | Well-differentiated |
| 7 | 75 | F | B | Rectosigmoid | Well-differentiated |
| 8 | 62 | M | C | Rectum | Well-differentiated |
| 9 | 71 | F | C | Sigmoid colon | Moderately-differentiated |
| 10 | 76 | M | B | Rectosigmoid | Well-differentiated |
| 12 | 70 | M | C | Sigmoid colon | Moderately-differentiated |
| 13 | 63 | M | B | Rectum | Moderately-differentiated |
| 14 | 62 | F | B | Rectosigmoid | Well-differentiated |
| 15 | 54 | F | B | Descending colon | Well-differentiated |
| 17 | 74 | M | B | Rectosigmoid | Moderately-differentiated |
| 19 | 67 | M | B | Rectum | Moderately-differentiated |
| 20 | 53 | F | C | Rectosigmoid | Moderately-differentiated |
| 21 | 79 | F | C | Rectosigmoid | Moderately-differentiated |
| 23 | 67 | M | C | Sigmoid colon | Moderately-differentiated |
| 24 | 42 | M | B | Descending colon | Well-differentiated |
The membrane or membrane-associated proteins were extracted from the homogenised tissues using thiourea buffer (7 mol/L urea, 2 mol/L thiourea, 4% CHAPS and 0.2% carrier ampholytes). Briefly, 200 mg of tissue was homogenised in 1 mL of Tris buffer (40 mmol/L Tris) followed by sonication for 30 s and chilled on ice for 2 min. The lysate was then vortexed for 5 min before being centrifuged (12 000 r/min, 15 min, 20°C). The supernatant was kept for a separate experiment while the pellet was rinsed twice with Tris buffer. The pellet was then suspended in 150 μL of thiourea buffer, sonicated, vortexed and centrifuged (12 000 r/min, 15 min, 20°C) and the supernatant was collected for analysis.
The protein concentration of the thiourea buffer extracts was determined using the RCDC protein assay kit (Bio-Rad, USA), and 500 μg of the extract in 185 μL of rehydration buffer (same composition as thiourea buffer) was used to rehydrate IPG strips (4-7 pH, 11 cm) for 15 h and focused using IEF Cell (Bio-Rad, USA) starting from 0 to 250 V within 15 min, followed by 250 to 8000 V within 2.5 h and maintained at 8000 V until 35 000 V-h was achieved. Subsequently, the IPG strips were equilibrated for 15 min with gentle shaking in Sodium Dodecyl Sulfate-Polyacrylamide Gel electrophoresis (SDS-PAGE) Equilibration Buffer I (6 mol/L urea, 0.375 mol/L Tris pH 8.8, 2% SDS, 20% glycerol, 2% DTT and a trace amount of bromophenol blue), followed by another 15 min of gentle shaking in SDS-PAGE Equilibration Buffer II (same composition as SDS-PAGE Equilibration Buffer 1, however, 2.5% iodoacetamide was used instead of 2% DTT). Second dimension separation was carried out under constant voltage of 200 V for approximately 3 h in 10% SDS-PAGE. The gel was stained overnight using Coomassie Blue (0.2% Coomassie Brilliant Blue R250, 50% MeOH and 2% acetic acid) and destained for 2 h.
The 2D-PAGE images were acquired by using the Versadoc system (Bio-Rad, USA). The gel images were processed and analyzed using PDQuest version 7.3 (Bio-Rad, USA). The software was used to create a matchset to compare the images of cancerous and normal colorectal tissues. The matchset was used to analyze quantitative and qualitative differences in protein spots between the images. The intensity of the protein spot was measured after normalization of the protein spot as a percentage of the total density of all proteins spots on each gel in order to minimise the variation which may be caused by the amount of sample loaded. β-actin protein spot was used as the landmark for gel image analysis. A protein was up-regulated if its expression level in cancerous tissues was 1.5-fold or more compared to normal colorectal tissue, and was down-regulated when the opposite occurred. A protein was uniquely expressed if it was found exclusively in either normal or cancerous colorectal tissue only. In addition, the statistical significance of the protein’s change in expression was determined using the Wilcoxon signed-rank test (PDQuest version 7.3) at 95% level of confidence.
In-gel digestion was carried out according to Gam et al[13]. In brief, the protein spots of interest were excised from the gel. The Coomassie blue stain on the protein spot was removed by dehydrating the gel pieces in acetonitrile and rehydrating in 100 mmol/L NH4HCO3. This step was repeated three times. The gel pieces were then incubated in a volume of trypsin buffer (50 mmol/L NH4HCO3 and 5 mmol/L CaCl2) containing 12.5 ng/μL trypsin and chilled at 4°C for 45 min. The trypsin buffer with trypsin was then replaced by trypsin buffer without trypsin at a volume sufficient to wet the gel pieces and incubated overnight at 37°C. The peptides were then eluted from the gel pieces and dried using a sample dryer (Techne, UK) under a continuous flow of nitrogen gas and stored at -20°C prior to analysis.
The dry peptides were suspended in 15 μL of ultrapure ddH2O and were subjected to liquid chromatography tandem mass spectrometry (LC/MS/MS) analysis using an electrospray ionisation-ion trap mass spectrometer (Agilent). A 5 μL volume of the reconstituted sample was injected into a RPC-column (C18, 300 Å, 5 μm, 1 mm × 150 mm) connected to a HPLC (1100 Series, Agilent). A capillary pump was used to pump the mobile phase (A and B) at a flow rate of 15 μL/min. The linear gradient used was 5% B to 95% B in 65 min. Mobile phase A was 0.05% formic acid in deionized water and B was 0.05% formic acid in acetonitrile. The HPLC was interfaced to the mass spectrometer detector. An experimental method comprising 2 scan events was used for analysis. The first scan event was a full scan MS whilst the second scan was the data dependent MS/MS scan which is dependent on the results of the first scan event. Two of the most intense ions in the MS scan which surpassed the threshold set were automatically isolated and excited to the MS/MS scan. The MS parameters used were; dry gas flow rate of 15 μL/min, nebulizer pressure of 30.0 psi and dry gas temperature of 300°C. The parameters for the MS/MS scan were; default collision energy (voltage) of 1.15 V, charge state of 2, minimum threshold of 1000 counts, and isolation width of 2 m/z. The MS/MS data from the analysis were used to search for their corresponding protein identity in Swiss-Prot using MASCOT Search engine version 2.2 from Matrix Science (http://www.matrixscience.com). The search parameters used were Homo sapiens for taxonomy, trypsin for enzyme, carboxymethyl for fixed modifications, peptide tolerance of +/- 2 Da, MS/MS tolerance of +/- 0.8 Da and average experimental mass value. Further analysis of proteins was carried out using the ProtParam programme available at the EXPASY website (http://www.expasy.org/tools/protparam.html) for calculation of the proteins grand average of hydrophobicity (GRAVY). The Tmpred programme (http://www.ch.embnet.org/software/TMPRED_form.html) was used to determine the transmembrane domain of the proteins.
Western blotting was carried out using a semi-dry blotting method[14]. Protein extracts were separated by SDS-PAGE according to Laemmli[15]. A similar quantity of protein was loaded on to SDS-PAGE, after electrophoresis separation, the proteins in the gel were transferred using a TE 70 Semiphor semi-dry transfer unit (Hoefer Scientific, Germany) at 134 mA for 1.5 h to a nitrocellulose membrane. The membrane was incubated in 20 mL of mouse anti-SLP-2 antibody (Abnova, Taiwan) at 1:250 followed by incubation with 50 mL of HRP conjugated anti-mouse secondary antibody (Bio-Rad, USA). The reaction of HRP and its substrate 4-Chloro Naphthol (4CN) indicated the presence of stomatin-like protein 2 (SLP-2).
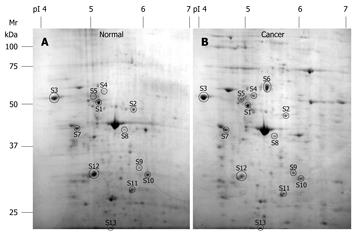
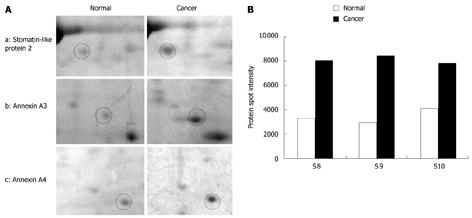
Two-dimensional gel electrophoresis for protein separation was carried out on a linear pH range of 4 to 7. A total of 13 differentially expressed proteins which were expressed abundantly in either cancerous or normal tissues were identified (Figure 1). Identification of differential protein expression in individual patients was accomplished by conducting a pair-wise comparison between the cancerous and normal tissues for each patient and is displayed in Figure 2. An average of 177.35 ± 26.60 protein spots was detected on 2D gel, with a coefficient variation of 15%. Eight proteins, namely tubulin α-1 chain (S4), tubulin β-2 chain (S5), chaperonin GroEL (S6), heat shock 70 kDa protein (S7), SLP-2 (S8), annexin A3 (S9), annexin A4 (S10) and ATP synthase D chain (S13) were up-regulated although only the up-regulation of tubulin β-2 chain, SLP-2, annexin A3 and annexin A4 were significant (P < 0.05) in CRC. Figure 3 shows the comparative analysis of spot intensity between normal and cancerous tissues for SLP-2, annexin A3 and annexin A4.
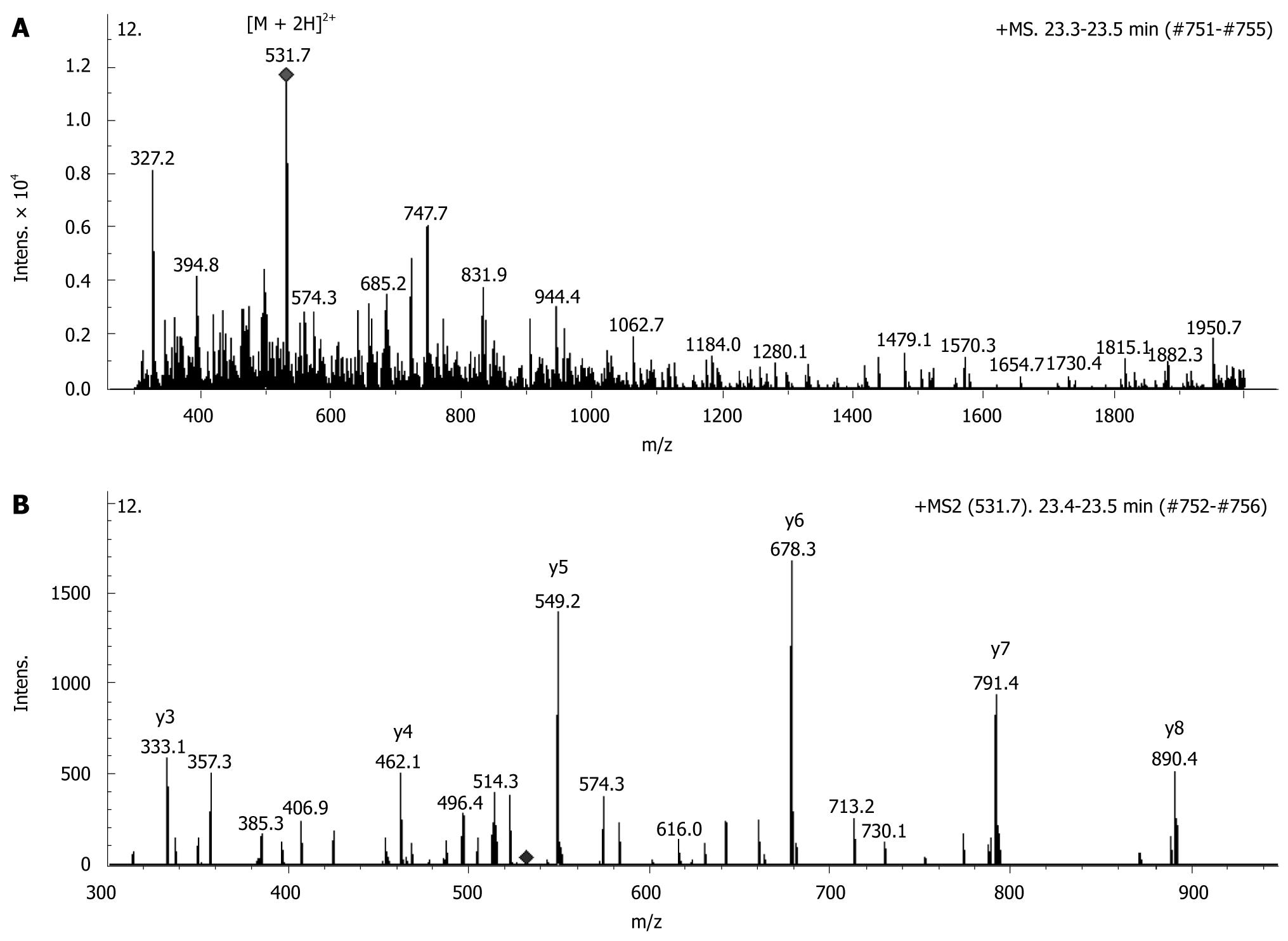
The identity of the proteins was determined by amino acid sequencing via tandem mass spectrometric analysis followed by protein database search. The representative MS and MS/MS spectra of SLP-2 are shown in Figure 4. Western blotting was used to confirm the results obtained from mass spectrometric analysis. Figure 5 shows a Western blot image of SLP-2 extracted from normal and cancerous tissue in the same patients, SLP-2 was only detected in cancerous tissue. Table 2 shows the 13 differentially expressed proteins identified in this study. The GRAVY score indicated the hydrophobic property of each protein, and the change in the protein expression levels is indicated as fold change (calculated as the ratio of total spot intensity of the protein in normal and cancerous tissues in all 18 patients). A positive value indicated that the protein expression level was higher in cancer compared to normal tissue or that it was up-regulated, while a negative value showed that the protein was down-regulated. Chaperonin GroEL was shown to have the greatest fold change (+265.0) although its up-regulation in all 18 patients was not statistically significant (P < 0.05).
| Protein spot | Protein name | SwissProt accession No. | Score | Theoretical molecular weight (Da) | Theoretical pI | Sequence coverage (%) | GRAVY | Fold change1 | TMR |
| S1 | F1-ATPase β subunit | P06576 | 364 | 58 013 | 5.80 | 19 | -0.030 | +58.5 | 1 |
| S2 | Ubiquinol-cytochrome c reductase | P31930 | 171 | 53 308 | 5.94 | 7 | -0.141 | +32.8 | 1 |
| S3 | Calreticulin | P27797 | 73 | 47 092 | 4.30 | 11 | -1.191 | -31.9 | 1 |
| S41f | Tubulin α-1 chain | Q71U36 | 61 | 50 800 | 4.94 | 6 | -0.229 | +10.8 | 1 |
| S51cf | Tubulin β-2 chain | P68371 | 299 | 48 142 | 4.70 | 25 | -0.347 | +20.8 | 1 |
| S6 | Chaperonin GroEL | P10809 | 185 | 61 348 | 5.70 | 8 | -0.076 | +265.0 | 3 |
| S7 | Heat shock 70 kDa protein | P11021 | 775 | 72 488 | 5.07 | 42 | -0.487 | +5.1 | 1 |
| S8abcdefg | Stomatin-like protein 2 | Q9UJZ1 | 151 | 38 644 | 6.88 | 28 | -0.161 | +27.9 | 1 |
| S9ae | Annexin A3 | P12429 | 140 | 36 396 | 5.63 | 22 | -0.430 | +34.9 | 0 |
| S10abcef | Annexin A4 | P09525 | 165 | 35 983 | 5.85 | 33 | -0.447 | +29.6 | 0 |
| S11 | Prohibitin | P35232 | 421 | 29 890 | 5.57 | 41 | +0.024 | +11.8 | 1 |
| S12 | Annexin A5 | P08758 | 195 | 35 994 | 4.94 | 39 | -0.330 | -5.2 | 0 |
| S13 | ATP Synthase D chain | O75947 | 117 | 18 406 | 5.22 | 32 | -0.569 | +8.3 | 0 |
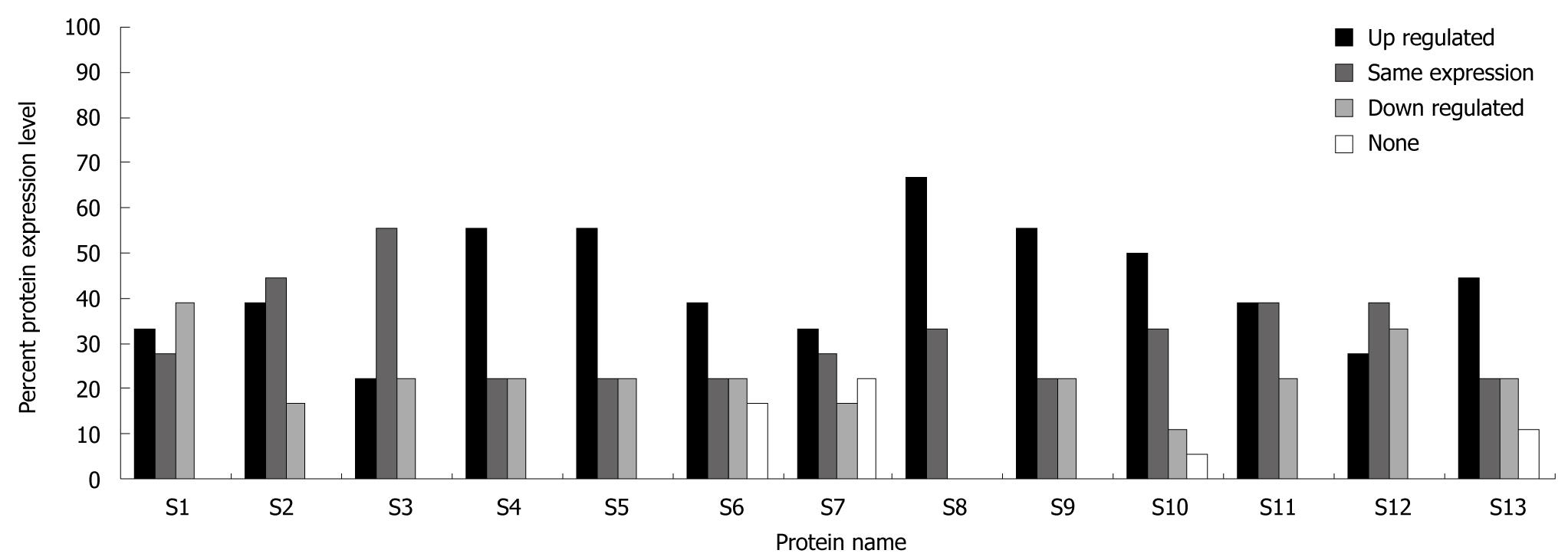
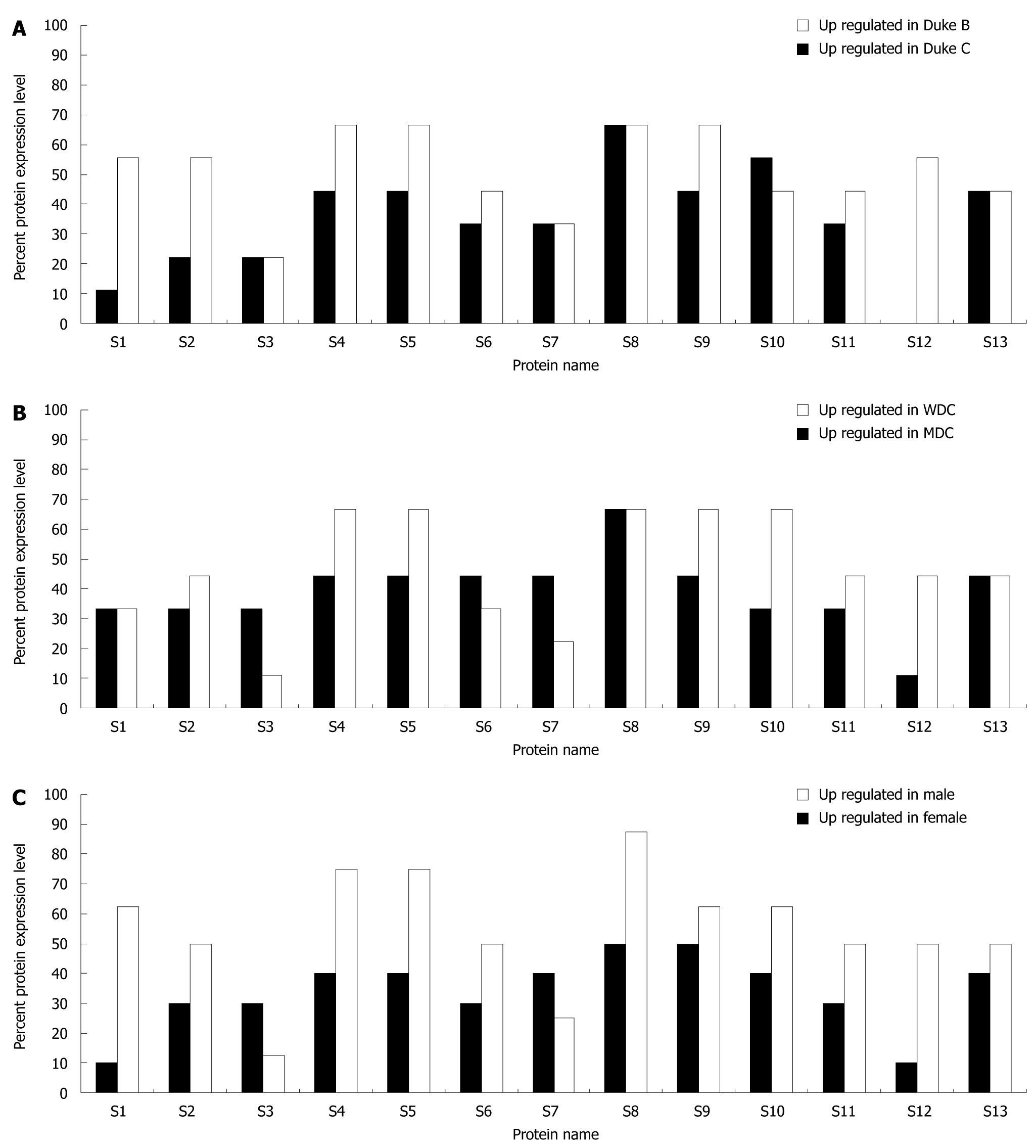
Analysis of protein expression was carried out by comparing protein expression profiles of cancerous and normal tissues within and between cancer stage, pathological status (grade) and gender of the patients. The tissue specimens collected comprised 9 each of Duke’s B and Duke’s C, respectively. Figure 6A shows the level of up-regulation of the 13 proteins in Duke’s B and C. The proteins which were up-regulated in > 50% of Duke’s B cancer were SLP-2 (S8) and annexin A4 (S10), while F1-ATPase β subunit (S1), uniquinol-cytochrome c reductase (S2), tubulin α-1 chain (S4), tubulin β-2 chain (S5), SLP-2 (S8), annexin A3 (S9) and ATP synthase D chain (S13) were up-regulated in > 50% of Duke’s C cancer. The up-regulation of both SLP-2 and annexin A4 was significant (P < 0.05) in Duke’s B cancer, whilst only tubulin β-2 chain and SLP-2 were significantly (P < 0.05) up regulated in Duke’s C cancer.
As for cancer grade, the tissues collected comprised 9 each of well-differentiated adenocarcinoma (WDC) and moderately-differentiated adenocarcinoma (MDC), respectively. Figure 6B shows the level of up-regulation of the 13 proteins in WDC and MDC. SLP-2 was expressed at > 50% in WDC and its up-regulation in WDC was significant (P < 0.05). Tubulin α-1 chain (S4), tubulin β-2 chain (S5), SLP-2 (S8), annexin A3 (S9) and annexin A4 (S10) were up-regulated in > 50% MDC, however, only the expression levels of SLP-2, annexin A3 and annexin A4 were significant (P < 0.05).
There were 10 male patients and 8 female patients included in this study. All 13 proteins identified in the study were equal to or > 50% up-regulated in female patients except for calreticulin (S3) and heat shock 70 kDa protein (S7) (Figure 6C). In contrast, all 13 proteins were up-regulated in < 50% of male patients except SLP- 2 (S8) and annexin A3 (S9) which were up-regulated in 50% of male patients (Figure 6C). Only SLP-2 was significantly up-regulated (P < 0.05) in CRC male patients, while tubulin α-1 chain, tubulin β-2 chain, SLP- 2 and annexin A4 were significantly up-regulated in CRC female patients.
The potential use of membrane proteins in drug targeted therapy and diagnosis of diseases is enormous, where many of the biomarkers for indication of diseases are of membrane origin[16]. The membrane or membrane-associated proteins contain potential antibody recognition sites that may provide identification of cancer development[6]. Although the extraction of membrane proteins is difficult, a combination of reagents can be used to enhance the solubility of membrane proteins. In this study, tissues were homogenised and treated with Tris buffer prior to thiourea buffer extraction. This pre-treatment removed aqueous soluble proteins and minimised proteolysis. Thus, the remaining tissue pellet contained mainly hydrophobic proteins which were then extracted using thiourea buffer. Thiourea buffer is usually used as soluble buffer for hydrophobic proteins[17]. Dowling et al[18] and Alvarez-Chaver et al[9] reported proteomic data of membrane proteins isolated from colorectal cancer by using Triton X-114. Although some common differentially expressed proteins were identified in our study and in the study by Alvarez-Chaver et al[9], there were also different types of proteins reported by both studies which indicated the crucial roles of reagents in determining the type of protein extracted.
Separation of proteins was carried out using 2-D gel electrophoresis. A similar quantity of proteins from cancerous and normal tissues was used in the analysis to ensure that the change in protein intensity was a factor of tissue type and not tissue weight, this is because the density of cancerous tissues is generally higher than that of normal tissues. A total of 500 μg of protein was shown to be an ideal load for 2-D gel separation as it allows the visualisation of minute proteins with good spot quality, while abundant proteins were kept under the saturation level when stained with Coomassie Blue.
In this study, only those proteins that were consistently expressed in all the tissues were targeted, we believe that this approach will minimise the identification of false-positive proteins that result from tissue heterogeneity and sample handling. In addition, the normalisation of protein spots during gel image analysis also serves as a control for the determination of differential protein expression levels. We observed a consistent pattern of proteomes for normal and cancerous tissues, respectively (Figure 1). Nevertheless, the expression levels of the proteins varied between patients, where no single protein was solely up-regulated or down-regulated in either cancerous or normal tissues in all patients; i.e. an un-regulated protein in one patient can be expressed as a down-regulated protein in another patient. This observation may support the phenomenon that one drug does not fit all and therefore, customized drugs have become the current trend for the treatment of cancer. Nevertheless, protein expression level was found to be more consistent when the tissues were analysed according to cancer stage, grade and gender of the patients. The association of protein expression with stage and grade of CRC and gender of the patients showed the potential of developing stage, grade or gender specific treatment for CRC patients.
Duke’s classification of tumor invasion has been proved to correlate with patient survival[19]. A greater number of proteins were found to be consistently up-regulated in Duke’s C tissues compared to Duke’s B tissues. In Duke’s B cancer, the tumor has not yet metastasized and therefore may still undergo the process of differentiation, nevertheless, when it advances to Duke’s C stage, where cancer has metastasized to the lymph nodes, the tumor may also reach a certain level of maturity and reveal a greater consistency in the protein expression profile. In contrast, Kwong et al[20] reported that changes in the number of highly expressed proteins do not correlate with the progression of colorectal cancer from Duke’s stage B to D.
Well-differentiated adenocarcinoma is less aggressive than moderately-differentiated adenocarcinoma, where the former has a greater resemblance to normal cells. We found that the pattern of protein expression in well-differentiated adenocarcinoma was closer to normal tissues, where only 1 protein was up-regulated in > 50% of well-differentiated tissues compared to 5 proteins in moderately-differentiated tissues.
With regard to the gender of patients, female patients showed a greater consistency in protein expression compared with male patients. In general, all 13 proteins identified had a much greater level of expression in female patients than in male patients except for heat shock 70 kDa protein and calreticulin. Calreticulin was identified as a down-regulated protein in this study. It has been reported that men have higher probabilities of developing polyps and tumors than women, although women are more likely to develop right-sided polyps and right-sided tumors than men[21], which may explain the different protein expression profiles in the two genders.
All 13 proteins identified in this study were among the highly expressed proteins in either cancerous or normal colorectal tissues. Of these proteins, SLP-2 has been shown to be a promising biomarker for CRC, particularly for female patients, where it was up-regulated in 87.5% of female patients, whereas in male patients, it was found to be up-regulated in 50% of patients. Moreover, in Duke’s B, Duke’s C, WDC and MDC, its up-regulation levels were 66.7%. Up-regulation of SLP-2 was statistically significant (P < 0.05) in both male and female patients and in the stages and grades of CRC tested. Its detection in CRC has not been reported, although it was reported to be up-regulated in other types of cancer. Over expression of SLP-2 was identified in human esophageal squamous cell carcinoma, lung cancer, laryngeal cancer, and endometrial adenocarcinoma which indicated that SLP-2 over expression is very common in cancer development. SLP-2 was associated with different stages of tumor progression from normal tissue to premalignant and malignant lesions of the esophagus[22]. High expression of SLP-2 was also attributed to advanced stages of breast cancer[23]. SLP-2 was reported to be involved in regulating cell growth and cell adhesion in human oesophageal squamous cell carcinoma[22]. SLP-2 plays an important role in sustaining T cell activation through the antigen receptor that is required for T cell differentiation during immunomodulation. It may also be involved in regulating ion channel conductance and/or the organisation of sphingolipid and cholesterol-rich lipid rafts[24].
The GRAVY score analysis of the thirteen proteins identified in this study showed that only prohibitin had a positive GRAVY score (0.024) indicating its hydrophobic nature. Nevertheless, Blonder et al[25] suggested that the GRAVY calculation does not reliably predict the hydrophobic nature of a protein. Therefore, the presence of the transmembrane domain of a protein is collectively used to predict the nature of a protein. Tmpred analysis of human SLP-2 (356 amino acids) has shown that it contains a single transmembrane domain, which putatively consists of 19 hydrophobic amino acids (amino acid 5-24). SLP-2 is a novel and unusual member of the stomatin gene superfamily. It is a peripheral membrane protein[24].
Besides SLP-2, other up-regulated proteins with significant expression were annexin A3, annexin A4 and tubulin β-2 chain. Annexin A3 and annexin A4 are members of the annexin family, a family of calcium-regulated phospholipid-binding proteins. Annexin A3 induces the migration and tube formation of vascular endothelial cells by inducing hypoxia-inducible factor-1 (HIF-1), which in turn causes the secretion of vascular endothelial growth factor (VEGF), an important factor in angiogenesis[26]. Annexin A4 forms complexes with protein kinase C, which has roles in cancer progression and was shown to be up-regulated in colorectal cancer[27]. Annexins have been reported to be involved in disease processes such as neoplasia. Furthermore, changes in the expression of annexins were linked with tumorigenesis[28]. Over expression of annexins in primary CRC increase significantly with advancing tumor stage, which suggests that annexins play a role in the progression and development of CRC[29]. Over expression of annexin A4 with advancing tumor stage was correlated with its role in promoting tumor cell migration. The distinct localization of annexin A4 in tumor cells was implicated to the loss of cell-to-cell adhesion and therefore increased tumor cell spread[30]. Our data showed that up-regulation of annexin A3 increased when the cancer stage advanced from Duke’s B to Duke’s C, while the opposite was observed for annexin A4. There have been no previous reports of annexin A3 participating in tumorigenesis or its association with different stages of CRC. Annexin A3 was reported to be up-regulated in colorectal tumor tissues and its cellular location was predominantly membrane-associated as revealed by immunohistochemistry assay[31]. Immunohistochemistry staining of annexin A3 in prostate cancer has shown its possible relationship with cancer grade[32]. In this study, we also observed that up-regulation of annexin A3 and annexin A4 in CRC increased when the cancer progressed from well-differentiated adenocarcinoma to moderately-differentiated adenocarcinoma.
The tubulin β-2 chain is one of the components of the cytoskeleton which plays a complex role in cells[33]. Significant differences in β-tubulin expression in polyps and invasive colon cancers indicates its possible roles in invasive cancer development[34]. A significant relationship between the expression of tubulins and stages of rectal cancer was suggested to be useful in identifying Dukes’ B and Duke’s C rectal cancer[34]. In this study, we found that the expression of tubulin β-2 chain increased significantly when the cancer advanced from Duke’s B to Duke’s C.
In conclusion, we have identified four hydrophobic proteins, namely SLP-2, tubulin β-2 chain, annexin A3 and annexin A4 which were abundantly expressed in cancerous tissues compared with normal tissues of the colon. Although a limited number of tissues were tested, the expression of these proteins in colorectal cancer was found to be significant suggesting that the possible use of these protein biomarkers in drug targeted therapy and in the diagnosis of colorectal cancer are worth further investigation.
Colorectal cancer (CRC) is the third most common cancer worldwide. In Malaysia, the incidence of CRC is increasing. An evaluation of the protein profile of CRC tissues may lead to an understanding of the changes in protein expression when cancer progresses. Proteomics is an emerging tool to study proteins and therefore can be used for the identification of potential biomarkers in the detection and treatment of CRC.
Membrane proteins comprise approximately 30% of the total human proteome. They are important component of cells and perform vital cellular functions. Due to their hydrophobic nature, many membrane proteins are difficult to extract and remain under-represented in protein profiles.
The authors have successfully extracted hydrophobic proteins from CRC patients. This group of proteins are differentially expressed and significantly correlated with stage and grade of cancer and gender of the patients. In their study, stomatin-like protein 2 (SLP-2) was significantly up-regulated in cancerous tissues. SLP-2 has not been previously reported in CRC.
Hydrophobic proteins are mainly located on the cell membrane. Therefore, they have the potential to be cell surface markers that can be used in drug targeted therapies for CRC.
A hydrophobic protein is a protein that contains a stretch of amino acids that have hydrophobic side chains. These amino acids are arranged so that the hydrophobic side chains are placed outside the protein in the three dimensional structure of the protein. This part of the protein will tend to embed itself in lipid structures such as cell membranes.
This manuscript describes the identification of a number of protein biomarkers that are up regulated in colorectal cancer. Membrane proteins from both cancer and normal mucosa were isolated using 2-D gel electrophoresis and subjected to LC/MS/MS analysis.
Peer reviewer: Dr. Kevin J Spring, PhD, Conjoint Gastroenterology Laboratory, The Queensland Institute of Medical Research, the Bancroft Centre, Room H07, PO Royal Brisbane Hospital, Herston, QLD 4029, Australia
S- Editor Wang YR L- Editor Webster JR E- Editor Ma WH
| 1. | World Health Organization. Cancer: Fact sheet. 2009 [cited 31th August 2009];. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/index.html. |
| 2. | Lim GCC, Yahaya H, Lim TO. The first report of the national cancer registry, cancer incidence in malaysia. Kuala Lumpur: Ministry of Health 2003; 57-58. |
| 3. | Alessandro R, Belluco C, Kohn EC. Proteomic approaches in colon cancer: promising tools for new cancer markers and drug target discovery. Clin Colorectal Cancer. 2005;4:396-402. |
| 4. | Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029-1038. |
| 5. | Tan S, Tan HT, Chung MC. Membrane proteins and membrane proteomics. Proteomics. 2008;8:3924-3932. |
| 6. | Canelle L, Bousquet J, Pionneau C, Hardouin J, Choquet-Kastylevsky G, Joubert-Caron R, Caron M. A proteomic approach to investigate potential biomarkers directed against membrane-associated breast cancer proteins. Electrophoresis. 2006;27:1609-1616. |
| 7. | Rabilloud T. Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis. 1998;19:758-760. |
| 8. | Rabilloud T. Solubilization of proteins in 2-D electrophoresis. An outline. Methods Mol Biol. 1999;112:9-19. |
| 9. | Alvarez-Chaver P, Rodríguez-Piñeiro AM, Rodríguez-Berrocal FJ, Martínez-Zorzano VS, Páez de la Cadena M. Identification of hydrophobic proteins as biomarker candidates for colorectal cancer. Int J Biochem Cell Biol. 2007;39:529-540. |
| 10. | Molloy MP. Two-dimensional electrophoresis of membrane proteins using immobilized pH gradients. Anal Biochem. 2000;280:1-10. |
| 11. | Dreger M. Subcellular proteomics. Mass Spectrom Rev. 2003;22:27-56. |
| 12. | Görg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665-3685. |
| 13. | Gam LH, Leow CH, Man CN, Gooi BH, Singh M. Analysis of differentially expressed proteins in cancerous and normal colonic tissues. World J Gastroenterol. 2006;12:4973-4980. |
| 14. | Laurière M. A semidry electroblotting system efficiently transfers both high- and low-molecular-weight proteins separated by SDS-PAGE. Anal Biochem. 1993;212:206-211. |
| 15. | Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680-685. |
| 16. | Lund R, Leth-Larsen R, Jensen ON, Ditzel HJ. Efficient isolation and quantitative proteomic analysis of cancer cell plasma membrane proteins for identification of metastasis-associated cell surface markers. J Proteome Res. 2009;8:3078-3090. |
| 17. | Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037-1053. |
| 18. | Dowling P, Meleady P, Dowd A, Henry M, Glynn S, Clynes M. Proteomic analysis of isolated membrane fractions from superinvasive cancer cells. Biochim Biophys Acta. 2007;1774:93-101. |
| 19. | Deans GT, Parks TG, Rowlands BJ, Spence RA. Prognostic factors in colorectal cancer. Br J Surg. 1992;79:608-613. |
| 20. | Kwong KY, Bloom GC, Yang I, Boulware D, Coppola D, Haseman J, Chen E, McGrath A, Makusky AJ, Taylor J. Synchronous global assessment of gene and protein expression in colorectal cancer progression. Genomics. 2005;86:142-158. |
| 21. | McCashland TM, Brand R, Lyden E, de Garmo P. Gender differences in colorectal polyps and tumors. Am J Gastroenterol. 2001;96:882-886. |
| 22. | Zhang L, Ding F, Cao W, Liu Z, Liu W, Yu Z, Wu Y, Li W, Li Y, Liu Z. Stomatin-like protein 2 is overexpressed in cancer and involved in regulating cell growth and cell adhesion in human esophageal squamous cell carcinoma. Clin Cancer Res. 2006;12:1639-1646. |
| 23. | Cao W, Zhang B, Liu Y, Li H, Zhang S, Fu L, Niu Y, Ning L, Cao X, Liu Z. High-level SLP-2 expression and HER-2/neu protein expression are associated with decreased breast cancer patient survival. Am J Clin Pathol. 2007;128:430-436. |
| 24. | Wang Y, Morrow JS. Identification and characterization of human SLP-2, a novel homologue of stomatin (band 7.2b) present in erythrocytes and other tissues. J Biol Chem. 2000;275:8062-8071. |
| 25. | Blonder J, Goshe MB, Moore RJ, Pasa-Tolic L, Masselon CD, Lipton MS, Smith RD. Enrichment of integral membrane proteins for proteomic analysis using liquid chromatography-tandem mass spectrometry. J Proteome Res. 2002;1:351-360. |
| 26. | Park JE, Lee DH, Lee JA, Park SG, Kim NS, Park BC, Cho S. Annexin A3 is a potential angiogenic mediator. Biochem Biophys Res Commun. 2005;337:1283-1287. |
| 27. | Gökmen-Polar Y, Murray NR, Velasco MA, Gatalica Z, Fields AP. Elevated protein kinase C betaII is an early promotive event in colon carcinogenesis. Cancer Res. 2001;61:1375-1381. |
| 28. | Rand JH. The annexinopathies: a new category of diseases. Biochim Biophys Acta. 2000;1498:169-173. |
| 29. | Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer. 2008;98:426-433. |
| 30. | Zimmermann U, Balabanov S, Giebel J, Teller S, Junker H, Schmoll D, Protzel C, Scharf C, Kleist B, Walther R. Increased expression and altered location of annexin IV in renal clear cell carcinoma: a possible role in tumour dissemination. Cancer Lett. 2004;209:111-118. |
| 31. | Madoz-Gúrpide J, López-Serra P, Martínez-Torrecuadrada JL, Sánchez L, Lombardía L, Casal JI. Proteomics-based validation of genomic data: applications in colorectal cancer diagnosis. Mol Cell Proteomics. 2006;5:1471-1483. |
| 32. | Wozny W, Schroer K, Schwall GP, Poznanović S, Stegmann W, Dietz K, Rogatsch H, Schaefer G, Huebl H, Klocker H. Differential radioactive quantification of protein abundance ratios between benign and malignant prostate tissues: cancer association of annexin A3. Proteomics. 2007;7:313-322. |
| 33. | Sullivan KF. Structure and utilization of tubulin isotypes. Annu Rev Cell Biol. 1988;4:687-716. |
| 34. | Giarnieri E, De Francesco GP, Carico E, Midiri G, Amanti C, Giacomelli L, Tucci G, Gidaro S, Stroppa I, Gidaro G. Alpha- and beta-tubulin expression in rectal cancer development. Anticancer Res. 2005;25:3237-3241. |