Published online Jul 7, 2009. doi: 10.3748/wjg.15.3178
Revised: May 25, 2009
Accepted: June 1, 2009
Published online: July 7, 2009
AIM: To examine whether 2'-5'oligoadenylate synthetase (OAS) gene promoter can be specifically activated by hepatitis C virus (HCV)-core protein.
METHODS: Human embryo hepatic cell line L02 was transfected with pcDNA3.1-core plasmid and selected by G418. Expression of HCV-core was detected by reverse transcription polymerase chain reaction and Western blotting. The OAS promoter sequence was amplified from the genomic DNA and inserted into pGL3-basic vector. The resultant pGL3-OAS-Luci plasmid was transiently transfected into L02/core cells and luciferase activity was assayed.
RESULTS: L02/core cell line stably expressing HCV-core protein was established. The pGL3-OAS-Luci construct exhibited significant transcriptional activity in the L02/core cells but not in the L02 cells.
CONCLUSION: HCV-core protein activates the OAS gene promoter specifically and effectively. Utilization of OAS gene promoter would be an ideal strategy for developing HCV-specific gene therapy.
- Citation: Wang Y, Mao SS, He QQ, Zi Y, Wen JF, Feng DY. Specific activation of 2'-5'oligoadenylate synthetase gene promoter by hepatitis C virus-core protein: A potential for developing hepatitis C virus targeting gene therapy. World J Gastroenterol 2009; 15(25): 3178-3182
- URL: https://www.wjgnet.com/1007-9327/full/v15/i25/3178.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.3178
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma worldwide[1]. Currently, there is no vaccine to prevent the infection and no specific antiviral drug directed against the disease. Gene therapy has emerged as a novel approach to combat HCV infection in the last few years[23]. However, one of the most important obstacles to overcome is “targeting”: the appropriate genes must be delivered to and expressed in HCV infected hepatocytes without harming normal tissues. This problem could be addressed by using a promoter that has a higher level of activity in HCV infected liver cells vs normal cells.
HCV is a member of the Flaviviridae family, containing approximately 9.5 kb of positive strand RNA. The viral genome encodes a large precursor polyprotein which is cleaved into functional proteins such as core, envelope (E1, E2) and non-structural proteins (NS2-NS5)[45]. The viral core protein consists of 191 amino acids and has an apparent molecular mass of 21 kDa. In addition to being the viral capsid protein, it also functions as a transcriptional regulator of various viral and cellular promoters. Some groups have reported that HCV-core protein activates the human c-myc, Erk, IL-2, and SV40 early promoters, and trans-suppresses some cellular promoters such as p53 and p21[6–12]. Though the mechanisms of these regulations are still unclear, it is possible that these properties of HCV-core protein influence host cell growth, survival, and carcinogenesis. Therefore, utilization of a promoter that is predominantly active in HCV-core positive hepatocytes would be an ideal strategy for developing HCV-specific gene therapy.
During the investigation of interferon (IFN)-inducible JAK-STAT signaling affected by HCV, Naganuma et al[13] found that HCV-core protein specifically activated the 2'-5'oligoadenylate synthetase (OAS) gene promoter in human hepatocyte cells, while the E1, E2, and NS5A proteins did not activate the OAS gene promoter. These data indicate that OAS promoter might be used to restrict gene expression in HCV-core positive liver cells. However, the role of HCV-core protein in modulating OAS gene expression is much controversial, some other groups have reported that HCV-core protein does not affect the activation of IFN-responsive genes (including OAS gene), although it modulates the JAK/STAT signaling pathway[14–16]. So whether this viral core protein interferes with the OAS promoter and whether the promoter could be used to target gene therapy remain to be demonstrated. For these reasons, in the present study, we cloned the OAS promoter and examined its activity in the HCV-core expressing liver cells in vitro.
Human embryo hepatocyte derived L02 cell line was obtained from Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences (Shanghai, China). The cells were routinely grown in DMEM medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 &mgr;g /mL streptomycin.
The pcDNA3.1-core, which contains the complete coding region of HCV-core protein (1b genotype) under the control of cytomegalovirus (CMV) immediate early promoter, was kindly provided by Professor Jun Cheng (Institute for Epidemic Disease Research, Beijing Ditan Hospital, China). The L02 cells were seeded in 6-well culture plates at 70% confluence 24 h before transfection. Cells were transfected with 2 &mgr;g of pcDNA3.1-core plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For establishment of stable cell line, the transfected cells were subjected to selection medium containing 400 mg/L G418 until several G418-resistant clones (L02/core) were obtained, and then the cells were maintained in G418 at a concentration of 200 mg/L. Non-transfected L02 cells were used as parallel control.
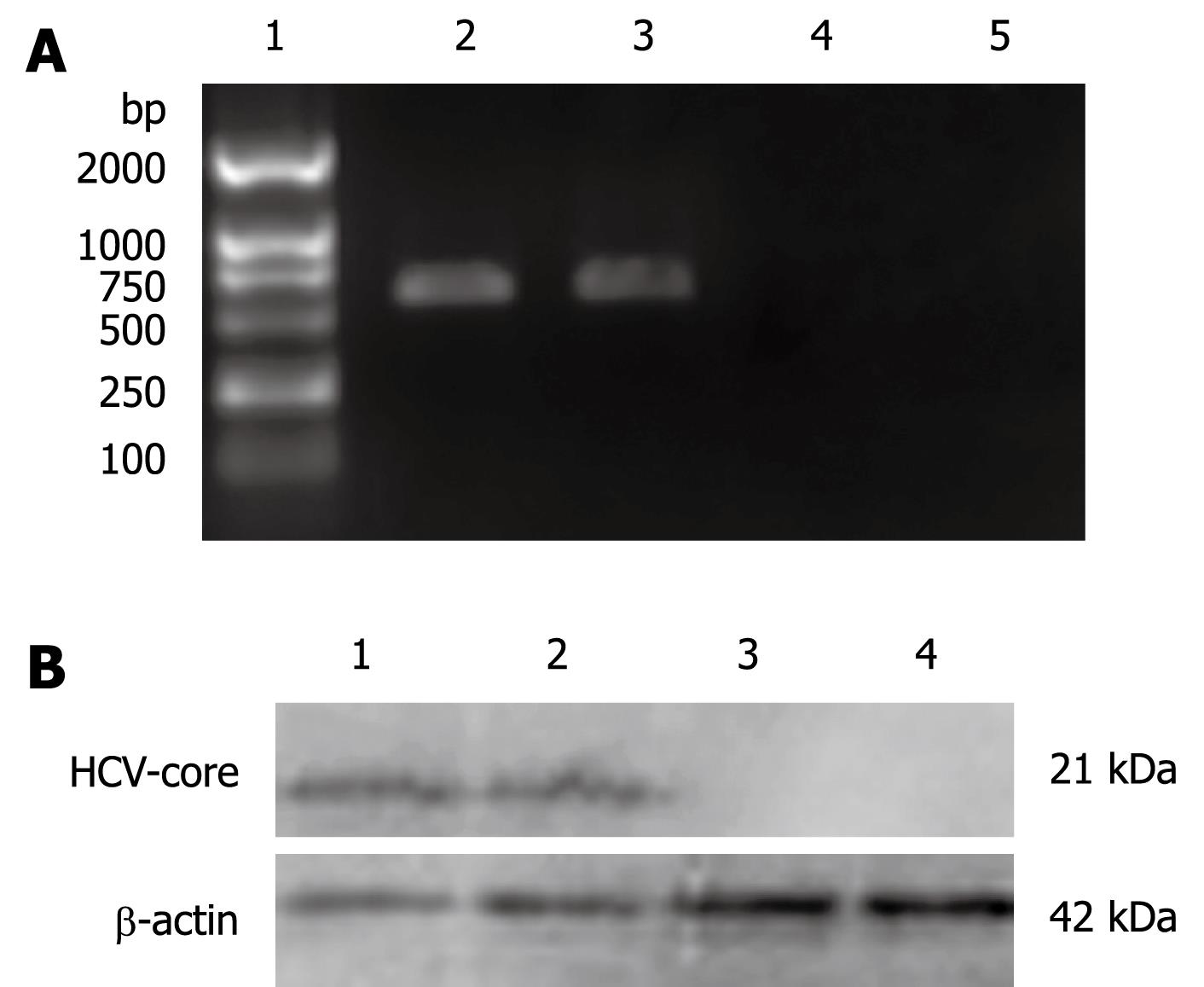
Total RNA was isolated from L02/core cells with TRIzol reagent (Invitrogen) and reversely transcribed using RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas). PCR was subsequently performed with HCV core-specific primers: 5'-ATGAGCACCAATCCTAAAC-3' (forward), and 5'-GGCTGAAGCGGGCACA-3' (reverse). The cycle parameters were as follows: 95°C for 1 min, 52°C for 50 s, and 72°C for 1 min; 30 cycles. The PCR products were electrophoresed in a 1.5% agarose gel containing ethidium bromide and visualized by UV illumination. The expected size of the PCR product for HCV-core mRNA was 572 base-pairs.
The L02/core cells were collected, washed with PBS and lyzed in ice-cold lysis buffer (20 mmol/L Tris HCl, pH 7.5, 150 mmol/L NaCl, 5 mmol/L EDTA, 1% Nonidet P-40, 1 mg/L Aprotinin, 100 mg/L PMSF). The protein concentration of the whole cell extract was measured using Bio-Rad DC protein assay kit. Equal amounts of cell lysates (60 &mgr;g protein) were separated by 12% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked for more than 1 h in PBS containing 2% nonfat milk and 0.25% Tween-20 and then immunoblotted using anti-HCV core monoclonal antibody (1:500, Santa Cruz). Immunoreactive bands were detected using horseradish-peroxidase-conjugated secondary antibodies (1:1000) in conjunction with an enhanced chemiluminescent system.
Template genomic DNA was extracted from human whole blood using DNAzol BD reagent (Invitrogen). The OAS1 promoter sequence (-157 to +82) was amplified by PCR from the genomic DNA. The primers incorporating SacI and HindIII restriction sites were: 5'-CCGAGCTCGGGATCAGGGGAGTGT-3' (forward) and 5'-CCCAAGCTTGCATGCGGAAACACG-3' (reverse). The PCR fragment was digested and cloned into the SacI/HindIII sites of pGL3-basic vector (Promega) to generate pGL3-OAS-Luci. Negative and positive control constructs were pGL3-basic lacking any promoter sequence, and pGL3-promoter containing the SV40 promoter sequence.
These luciferase reporter plasmids were transiently transfected into L02/core and L02 cells using Lipofectamine 2000 (Invitrogen). The cells were harvested 48 h after the transfection. Luciferase assays were performed according to the manufacturer’s protocols (Promega). Briefly, cells were lysed with reporter lysis buffer, and the luciferase activity was determined using a luminometer. A β-galactosidase expression plasmid (pSV-β-galactosidase; Promega) was co-transfected to allow normalization for transfection efficiency. All experiments were performed at least three times in each plasmid and represent the relative luciferase activity as average.
All data are shown as the mean ± SD. Statistical analysis was performed using the t test. P < 0.05 was considered statistically significant.
In order to investigate the effect of HCV-core on the OAS promoter, L02 cells were transfected with pcDNA3.1-core plasmid and were selected on the basis of their resistance to G418 for 2 wk. Expression of HCV-core was detected by RT-PCR and Western blotting. As shown in Figure 1A, 572 bp visible fragments, consistent with the predicted size of HCV-core mRNA, were detected in L02/core cell clones. No PCR-amplification product was found in the pcDNA3.1 transfected L02 cells, neither in the nontransfected L02 cells. The expression of HCV-core protein was confirmed by immunoblotting using anti-HCV core monoclonal antibody. As shown in Figure 1B, HCV-core protein of the expected molecular mass of 21 kDa was observed in L02/core cells, whereas no expression was detected in pcDNA3.1 transfected or nontransfected L02 cells. These data demonstrate that hepatocyte line stably expressing HCV-core protein has been established.
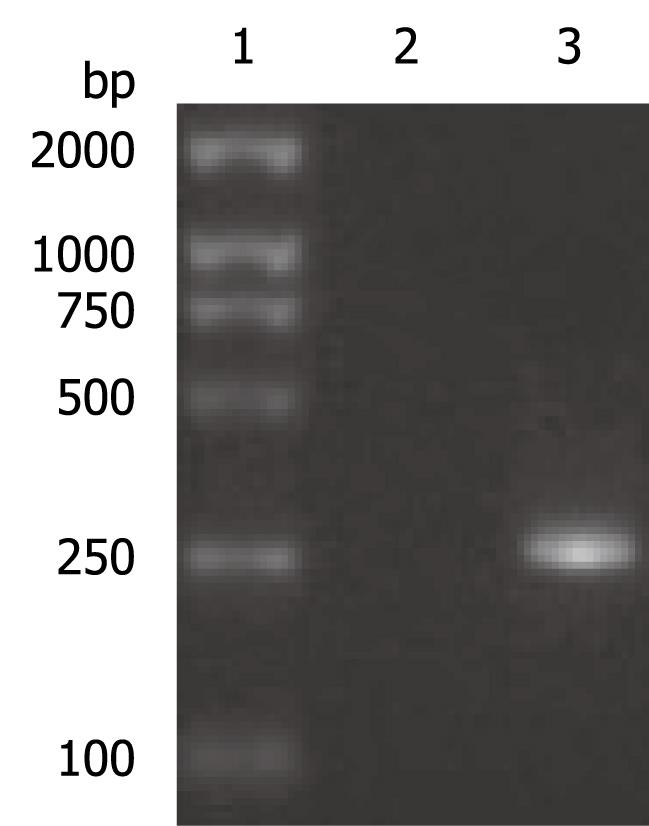
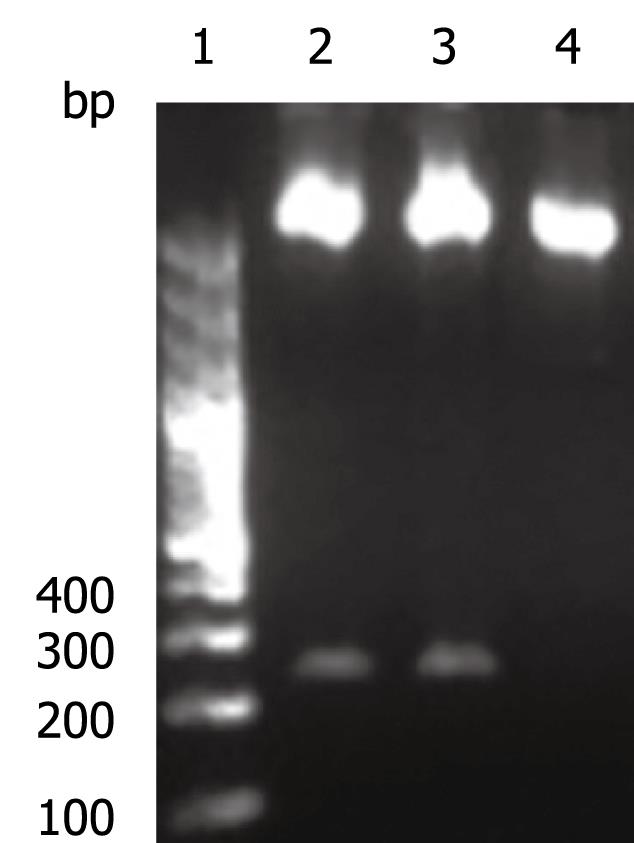
The OAS promoter sequence was amplified by PCR from human genomic DNA. As shown in Figure 2, a 240 bp visible fragment consistent with the predicted size was obtained by electrophoresis. Sequencing analysis confirmed that the cloned gene was identical with the original sequence in GenBank (accession number NW_925395). The OAS promoter was put upstream of the firefly luciferase gene in the pGL3-basic vector which lacks eukaryotic promoter and enhancer sequences. The resultant pGL3-OAS-Luci plasmid was confirmed by double enzyme digestion with SacI and HindIII (Figure 3).
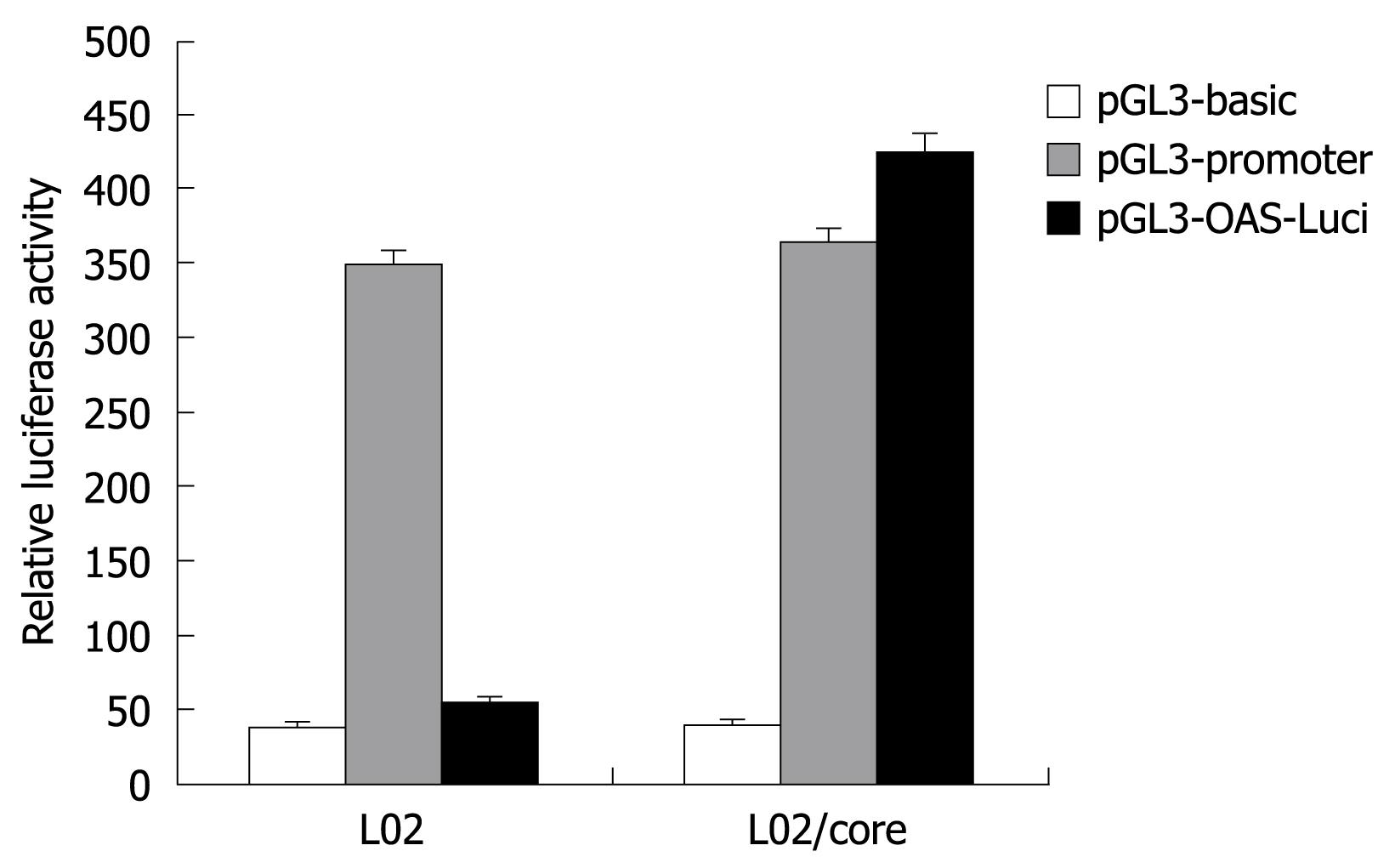
To examine the transcriptional activity of the OAS gene promoter, the pGL3-OAS-Luci was transiently transfected into L02/core and L02 cells, and the firefly luciferase activity obtained was compared with those from the pGL3-basic and pGL3-promoter plasmid. As shown in Figure 4, the pGL3-OAS-Luci construct exhibited significantly higher transcriptional activity in the presence of HCV-core protein (L02/core). The relative luciferase activity was almost 1.2 fold of pGL3-Promoter, and 10-15 fold of the baseline activity of the pGL3-basic vector. In contrast, this OAS promoter construct resulted in no or little luciferase activity in the normal liver cell line L02. These findings suggest that HCV-core protein strongly activates the OAS gene promoter.
To increase the specificity and safety of gene therapy, the expression of the therapeutic gene need to be tightly controlled within the target tissue. This is particularly important for toxic gene strategies because inappropriate transgene expression may lead to severe toxicity. Targeted expression of therapeutic genes can be accomplished at several levels. The first approach for targeting specificity is at the level of vector delivery[17]. The strategies include exploiting natural viral tropisms, and incorporating tissue-specific ligands or monoclonal antibodies onto the surface of viral vectors or liposomes. However, there will undoubtedly be some genes delivered to local and distant normal tissues. Therefore, further safeguards must be put in place to ensure that gene delivery to these tissues does not result in significant expression and toxicity. One attractive approach to this problem is to use promoter elements to control gene expression tightly at the transcriptional level.
Many tissue and tumor specific promoters have been developed in target gene therapy. For example, the α-fetoprotein promoter has been used to drive gene expression in hepatic carcinoma cells, the tyrosinase promoter in melanoma cells, the prostate specific antigen promoter in prostate cancer cells, and the carcinoembryonic antigen promoter in adenocarcinomas[18]. The results of these studies have demonstrated the feasibility of using specific promoters for targeting gene therapy in various cancer cell types.
Although several promoters have been identified more active in HCV-core positive hepatocytes, most of these promoters are much weaker than commonly used viral promoters such as the CMV early promoter, the Rous sarcoma virus long terminal repeat, and the SV40 early promoter. Consequently, their applications in gene therapy are hampered by the low expression. In the present study, we cloned the human OAS promoter and examined its transcriptional ability in the HCV-core positive hepatocytes and normal liver cells. We found that the luciferase expression driven by OAS promoter was markedly increased in the presence of core protein, but not in the normal liver cells. These data strongly suggest that HCV-core protein can activate OAS promoter. Since HCV-core protein plays an important role in persistent infection and hepatocellular carcinogenesis, and amino acid sequence of core protein is relatively conserved, utilization of the OAS promoter to drive therapeutic gene expression would be an ideal strategy for developing HCV-specific gene therapy.
The current standard therapy for chronic hepatitis C is inadequate for the majority of patients. Gene therapy has emerged as a novel approach to combat hepatitis C virus (HCV) infection in the last few years. However, a specific promoter is required to restrict transgene expression only in HCV infected cells. Since HCV-core protein plays an important role in persistent infection and hepatocellular carcinogenesis, and amino acid sequence of core protein is relatively conserved, utilization of a promoter that is predominantly active in HCV-core positive hepatocytes would be an ideal strategy for HCV targeting gene therapy.
To increase the specificity and safety of gene therapy, the expression of the therapeutic gene need to be tightly controlled within the target tissue. This is particularly important for toxic gene strategies because inappropriate transgene expression may lead to severe toxicity. Targeted expression of therapeutic genes can be accomplished at several levels, including vector targeting and tissue-specific gene expression. However, no HCV-specific gene delivery system has yet been developed. One attractive approach to this problem is to use promoter elements to control gene expression tightly at the transcriptional level.
In the present study, the authors cloned the 2'-5'oligoadenylate synthetase (OAS) promoter and examined its activity in the human embryo hepatocytes expressing HCV-core in vitro. They demonstrated that HCV-core protein can activate OAS gene promoter specifically and effectively.
Utilization of OAS gene promoter to drive therapeutic gene expression would be an ideal strategy for developing HCV-specific gene therapy.
L02/core is the human embryo hepatocyte that stably expresses HCV-core protein. OAS is a metabolic enzyme originally identified as a regulator of the ribonuclease L (RNase L) pathway during viral infection.
This is a well written and interesting paper.
| 1. | Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S-20S. |
| 2. | Gonzalez-Aseguinolaza G, Crettaz J, Ochoa L, Otano I, Aldabe R, Paneda A. Gene therapy for viral hepatitis. Expert Opin Biol Ther. 2006;6:1263-1278. |
| 3. | Ji J, Glaser A, Wernli M, Berke JM, Moradpour D, Erb P. Suppression of short interfering RNA-mediated gene silencing by the structural proteins of hepatitis C virus. J Gen Virol. 2008;89:2761-2766. |
| 4. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. |
| 5. | Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527-1538. |
| 6. | Ray RB, Lagging LM, Meyer K, Steele R, Ray R. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 1995;37:209-220. |
| 7. | Tsuchihara K, Hijikata M, Fukuda K, Kuroki T, Yamamoto N, Shimotohno K. Hepatitis C virus core protein regulates cell growth and signal transduction pathway transmitting growth stimuli. Virology. 1999;258:100-107. |
| 8. | Shrivastava A, Manna SK, Ray R, Aggarwal BB. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722-9728. |
| 9. | Kato N, Yoshida H, Ono-Nita SK, Kato J, Goto T, Otsuka M, Lan K, Matsushima K, Shiratori Y, Omata M. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology. 2000;32:405-412. |
| 10. | Bergqvist A, Rice CM. Transcriptional activation of the interleukin-2 promoter by hepatitis C virus core protein. J Virol. 2001;75:772-781. |
| 11. | Ray RB, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983-10986. |
| 12. | Lee MN, Jung EY, Kwun HJ, Jun HK, Yu DY, Choi YH, Jang KL. Hepatitis C virus core protein represses the p21 promoter through inhibition of a TGF-beta pathway. J Gen Virol. 2002;83:2145-2151. |
| 13. | Naganuma A, Nozaki A, Tanaka T, Sugiyama K, Takagi H, Mori M, Shimotohno K, Kato N. Activation of the interferon-inducible 2'-5'-oligoadenylate synthetase gene by hepatitis C virus core protein. J Virol. 2000;74:8744-8750. |
| 14. | Basu A, Meyer K, Ray RB, Ray R. Hepatitis C virus core protein modulates the interferon-induced transacting factors of Jak/Stat signaling pathway but does not affect the activation of downstream IRF-1 or 561 gene. Virology. 2001;288:379-390. |
| 15. | de Lucas S, Bartolome J, Carreno V. Hepatitis C virus core protein down-regulates transcription of interferon-induced antiviral genes. J Infect Dis. 2005;191:93-99. |
| 16. | Weber F. Interaction of hepatitis C virus with the type I interferon system. World J Gastroenterol. 2007;13:4818-4823. |
| 17. | Galanis E, Vile R, Russell SJ. Delivery systems intended for in vivo gene therapy of cancer: targeting and replication competent viral vectors. Crit Rev Oncol Hematol. 2001;38:177-192. |
| 18. | Robson T, Hirst DG. Transcriptional Targeting in Cancer Gene Therapy. J Biomed Biotechnol. 2003;2003:110-137. |