Published online May 7, 2009. doi: 10.3748/wjg.15.2089
Revised: February 19, 2009
Accepted: February 26, 2009
Published online: May 7, 2009
AIM: To analyze the relevance of the microRNA miR-196a for colorectal oncogenesis.
METHODS: The impact of miR-196a on the restriction targets HoxA7, HoxB8, HoxC8 and HoxD8 was analyzed by reverse transcription polymerase chain reaction (RT-PCR) after transient transfection of SW480 cancer cells. The miR-196a transcription profile in colorectal cancer samples, mucosa samples and diverse cancer cell lines was quantified by RT-PCR. Transiently miR-196a-transfected colorectal cancer cells were used for diverse functional assays in vitro and for a xenograft lung metastasis model in vivo.
RESULTS: HoxA7, HoxB8, HoxC8 and HoxD8 were restricted by miR-196a in a dose-dependent and gene-specific manner. High levels of miR-196a activated the AKT signaling pathway as indicated by increased phosphorylation of AKT. In addition, high levels of miR-196a promoted cancer cell detachment, migration, invasion and chemosensitivity towards platin derivatives but did not impact on proliferation or apoptosis. Furthermore, miR-196a increased the development of lung metastases in mice after tail vein injection.
CONCLUSION: miR-196a exerts a pro-oncogenic influence in colorectal cancer.
-
Citation: Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High
miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol 2009; 15(17): 2089-2096 - URL: https://www.wjgnet.com/1007-9327/full/v15/i17/2089.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.2089
Survival in colorectal cancer (CRC), one of the three most prevalent malignancies in western countries, is delineated by local recurrence, lymphatic and distant dissemination[1–3]. Molecular determinants occurring during the adenoma-carcinoma sequence of sporadic CRC include mutations in certain tumor-suppressor genes (APC, DCC, Smad-2, Smad-4, p53) and oncogenes (K-ras) that have been summarized by Fearon and Vogelstein[4–6]. However, as only 8% of CRCs harbor concomitant mutations of APC, K-ras and p53, it seems very likely that additional pathogenic alterations are instrumental in promoting progression and metastasis of colorectal cancer[7].
A recently discovered class of non-protein-coding small RNAs, microRNAs (miRNAs), extend our understanding of oncogenesis. miRNAs are endogenous small RNA molecules of 20-25 nucleotides length, regulating gene expression by inhibiting transcription, inducing direct cleavage of the targeted mRNAs or blocking translation through their complementarity versus targeted mRNAs at 3′ untranslated regions[8–13].
More than 50% of all known miRNA genes are located in cancer-associated regions or in fragile sites of the genome, indicating that miRNAs might play an important role in oncogenesis[14]. Supporting evidence is the close location of miRNAs, as miR-196a, in homeobox (Hox) gene clusters[14]. Hox proteins are major transcription factors that play a crucial role during embryogenesis, organogenesis and oncogenesis[15].
While some miRNAs can function as oncogenes, others act as tumor suppressors. Specific miRNAs, such as let-7, are under-expressed in cancer and function as tumor suppressors by regulating oncogenes in normal tissue. New evidence indicates that down-regulation of let-7 transcription is a relevant step during oncogenesis which is significantly associated with shortened postoperative survival in lung cancer[16–18]. Let-7 negatively regulates the expression of oncogenes Ras and Myc by targeting their mRNAs for translational repression in diverse malignancies[19].
In contrast, over-expressed miRNAs, such as miR-17-92, function as oncogenes promoting cancer development through inhibition of tumor suppressor genes. The expression of miRNA miR-17-92 is significantly increased in small-cell lung cancer[20]. Interestingly, the known targets of miR-17-92 include the two well-known tumor suppressor genes, PTEN and RB2[21].
The miRNA miR-196a, encoded at three locations in the mammalian Hox clusters A, B, and C, depicts evolutionarily conserved complementarity to mRNA of HoxB8, HoxC8, and HoxD8[22]. Interestingly, miR-196a-directed cleavage of HoxB8 was detected in mouse embryos, and additional in vivo experiments revealed a down-regulation of HoxB8, HoxC8, HoxD8 and HoxA7 in mammalian cells. These results indicate a miRNA-mediated regulation of Hox gene expression during vertebrate embryogenesis[22].
Matching these observations, Hornstein and colleagues describe that miR-196a acts upstream of HoxB8 and sonic hedgehog (Shh) in vivo during limb development[23]. Analyzing the miRNA expression pattern in pancreatic adenocarcinoma by large-scale miRNA chip analyses, Croce and colleagues found that 75% of tumors expressed miR-196a at a high level, predicting poor patient survival and linking miR-196a to human oncogenesis (14.3 mo vs 26.5 mo)[24].
As we had previously investigated the relevance of Hox genes for gastrointestinal cancer progression and observed a tumor-suppressive function of high HoxC8 expression levels, we hypothesized that miR-196a might exert a pro-oncogenic influence in human cancer cells.
The human colorectal cancer cell lines SW480, SW620 and HT29 and the human gastric cancer cell line Snu16 were cultured in RPMI-1640 (Invitrogen, Germany) supplemented with 10% FCS, 100 U/mL penicillin, 100 &mgr;g/mL streptomycin (Cambrex, Germany) and 1 mmol/L L-glutamine (Invitrogen, Germany).
Colorectal cancer and mucosal tissue has been collected from the resectate of seven patients undergoing elective surgery for colorectal cancer after obtaining patients’ written informed consent and approval by the local ethics committee.
miRNA isolation was performed from four cancer cell lines, and from seven colorectal cancer and matching mucosal samples using the MirVana miRNA Isolation Kit according to the manufacturer’s recommendations (Ambion, Austin, USA). HSA-miR-196a and U6 primer sets were commercially acquired and applied for quantitative RT-PCR using the MirVana QRT-PCR miRNA Detection Kit with Super Taq Polymerase (Ambion). For amplification, an Applied Biosystems 7900 HT Fast Realtime PCR System (Applied Biosystems, Foster City, USA) was used.
3 × 105 SW480 colon cancer cells were plated in a six-well plate and cultured as described before. SW480 cells were used, as they had the lowest miR-196a transcription levels (see below). miR-196a was commercially synthesized (MWG Biotech, Germany) and applied at different concentrations (0, 20, 40, 80, 160 and 240 nmol/L). Transfection was performed with Lipofectamine siRNAmax (Invitrogen, Carlsbad, CA, USA) according to the recommendations of the manufacturer. Cells were harvested 24-48 h after transfection and either applied in the functional assays, in a xenograft bioassay or collected for RNA/protein extraction, respectively.
6 × 103 transiently transfected SW480 cells (mock or 160 nmol/L miR-196a) were plated in 96-well plates and cultured as described above. The start of analyses was 24 h after transient transfection. The number of cells per well was determined daily by absorbance (MTT). Absorbance was quantified with an ELISA reader. Each condition was performed in quadruplicate.
For adhesion assays, SW480 cells were used. Transient transfection (mock or 160 nmol/L miR-196a) was performed 48 h prior to assay start. Ninety-six-well plates had been prepared with laminin (10 &mgr;g/mL, 30 min, room temperature, Sigma, Germany), fibronectin (10 &mgr;g/mL, 30 min, room temperature, Sigma) or PBS and were blocked with albumin (2%, overnight, 4°C, Serva, Germany), respectively. After trypsinization, 4 × 104 cells were seeded per 96-well and allowed to attach for 45 min. Thereafter, the medium and non-attached cells were removed. Each well was washed twice with 100 &mgr;L pure RPMI-1640 cell culture medium. The number of attached cells per well was determined by luminescence assay (Celltiter-Glo Cell Viability assay; Promega, USA). Emitted luminescence was quantified with a luminometer. Each condition was performed in quadruplicate. For dose-dependent quantification of adhesion (0, 40, 80 or 160 nmol/L miR-196a) non-modified 96-well plates were used.
For migration and invasion assays SW480 cells were used 48 h after transient transfection (mock or 160 nmol/L miR-196a). Migration and invasion were assayed with 24-well HTS FluoroBlock Inserts in triplet approaches (8 &mgr;mol/L pore size; Becton Dickinson, USA). For invasion assays, membranes were covered with fibronectin in advance (10 &mgr;g/mL, 30 min, room temperature, Sigma) and blocked with albumin (2%, overnight, 4°C, Serva).
In brief, 4 × 104 cells were re-suspended in serum-free RPMI-1640 medium and added to the upper chamber. Consecutively, RPMI-1640 medium with 20% FCS and 100 ng/mL CXCL12 was added to the lower chamber. Chambers were incubated for 24 h at 37°C in a humid atmosphere of 5% CO2. After incubation, the amount of cell invasion and migration into the lower chamber was determined by luminescence assay (Celltiter-Glo, Cell Viability assay; Promega) according to the recommendations of the manufacturers. Emitted luminescence was quantified with a luminometer. Each condition was performed in triplicate.
3 × 105 SW480 cells (mock or 160 nmol/L miR-196a) were seeded per six-well plate. Twenty-four hours after plating, 5-fluorouracil (5-FU) (10 &mgr;g/mL), irinotecan (40 &mgr;g/mL), oxaliplatin (10 &mgr;g/mL), cisplatin (20 &mgr;g/mL) or placebo (1 × PBS) were added to the medium. The number of apoptotic cells was determined after 48 h by apoptosis assay. In brief, suspension cells were collected and adherent cells were trypsinized prior to fixation with 100% ethanol, stained with propidium iodide and analyzed by FACS without gating. Each condition was performed in quadruplicate.
SW480 cells were harvested 2 d after transient transfection (mock or 160 nmol/L miR-196a), washed twice with PBS (1 ×) and lysed in 2 × RIPA solution. For Western blotting analysis, 100 µg of protein was loaded on a 13% SDS-PAGE gel. After separation, the gel was transferred to a PVDF membrane (Roth, Karlsruhe, Germany). AKT protein was detected with a rabbit-anti-human antibody (1:1000, overnight, 4°C, rabbit-anti-human monoclonal antibody, pan AKT, 4685; Cell Signaling, Danvers, MA, USA). Phosphorylated AKT (pAKT) protein was detected with a rabbit-anti-human antibody (1:1000, overnight, 4°C, rabbit-anti-human monoclonal antibody, Phospho-AKT, 9267, Cell Signaling). MEK1/2 was detected with a monoclonal rabbit-anti-human antibody (1:1000, overnight, 4°C, rabbit-anti-human monoclonal antibody, 9122; Cell Signaling). pMEK1/2 was detected with a monoclonal rabbit-anti-human antibody (1:1000, overnight, 4°C; rabbit-anti-human monoclonal antibody, 9121; Cell Signaling). Alpha-tubulin was analyzed with a monoclonal mouse-anti-human antibody (T5168, 1:2000, overnight, 4°C, Sigma). The secondary antibodies used were goat-anti-rabbit (1:10 000, 1 h, RT, SC-2033, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and goat-anti-mouse (1:10 000, 1 h, RT, SC-2031, Santa Cruz Biotechnology). For visualisation the Roti Lumin systems 1 and 2 were applied (P79 and P80; Roth).
Transient transfection (mock or 160 nmol/L miR-196a) of SW480 was performed 48 h prior to assay start. 4 × 104 tumor cells were re-suspended in 0.2 mL pure RPMI-1640 medium and applied for induction of lung metastases in 7-8-wk-old nod-Scid mice. Nod-Scid mice were radiated with 1.8 Gy 1 d prior to intravenous injection (tail vein) of tumor cells. Lung tumors grew for 7 wk before the animals were sacrificed. Thereafter, lungs were resected and tumor nodules quantified manually using surgical magnifying glasses.
RNA isolation was performed using the Qiagen RNeasy Kit according to the manufacturers recommendations (Qiagen, Hilden, Germany). Gene transcription of β-actin, HoxA7, HoxB8, HoxC8, HoxD8 was analyzed by a two-step RT-PCR: reverse transcription was performed with 2 &mgr;g of RNA (20 &mgr;L total volume; Ominscript RT Kit; Qiagen) according to the recommendations of the manufacturer. One microliter of cDNA was used as a template for the specific PCR reactions. Primers applied were β-actin-forward: 5'-TGACGGGGTCACCCACACTGTGCCCATCTA-3', β-actin-reverse: 5'-CTAGAAGCATTTGCGGTGGACGACGGAGGG-3' (661 bp fragment), HoxA7-forward: 5'-CCGCATGAAGTGGAAGAAAG-3', HoxA7-reverse: 5'-CAGTCCACAAAAGTTGGGAG-3' (347 bp fragment), HoxB8-forward: 5'-GCAATTTCTACGGCTACGAC-3' and HoxB8-reverse: 5'-GAAACAGAAGCTGGAGCGG-3' (434 bp fragment), HoxC8-forward: 5'-CACGTTCAAGACTTCTTCCACCACG-3' and HoxC8-reverse: 5'-GGTTCCAGAACCGAAGGATGAAGTG-3' (449 bp fragment), HoxD8-forward: 5'-ACAGCCGATTTTTACGACCC-3' and HoxD8-reverse: 5'-GCTTCCTTTTTCGTTTCCCC-3' (399 bp fragment).
For amplification, a DNA Engine PTC200 (MJ Research, Watertown, USA) thermocycler was used. Cycling conditions of the respective PCR were as follows: initial denaturation (4 min at 95°C), followed by the respective number of cycles (β-actin: 20; HoxA7: 29, HoxB8: 29, HoxC8: 29, HoxD8: 29) of denaturation (1 min at 94°C), annealing (1 min; β-actin: 57°C; HoxA7: 58°C, HoxB8: 56°C, HoxC8: 62°C, HoxD8: 57°C) and elongation (2 min at 72°C). After the last cycle, a final extension (10 min at 72°C) was added and thereafter the samples were kept at 4°C. Seven microliters of the products were run on a 1.8% agarose gel, stained by ethidium bromide and analyzed under UV light.
The χ2 test was used to compare all other patient and tumor characteristics by group. The t test was applied to compare results obtained from function assays. For all tests, P < 0.05 was considered significant.
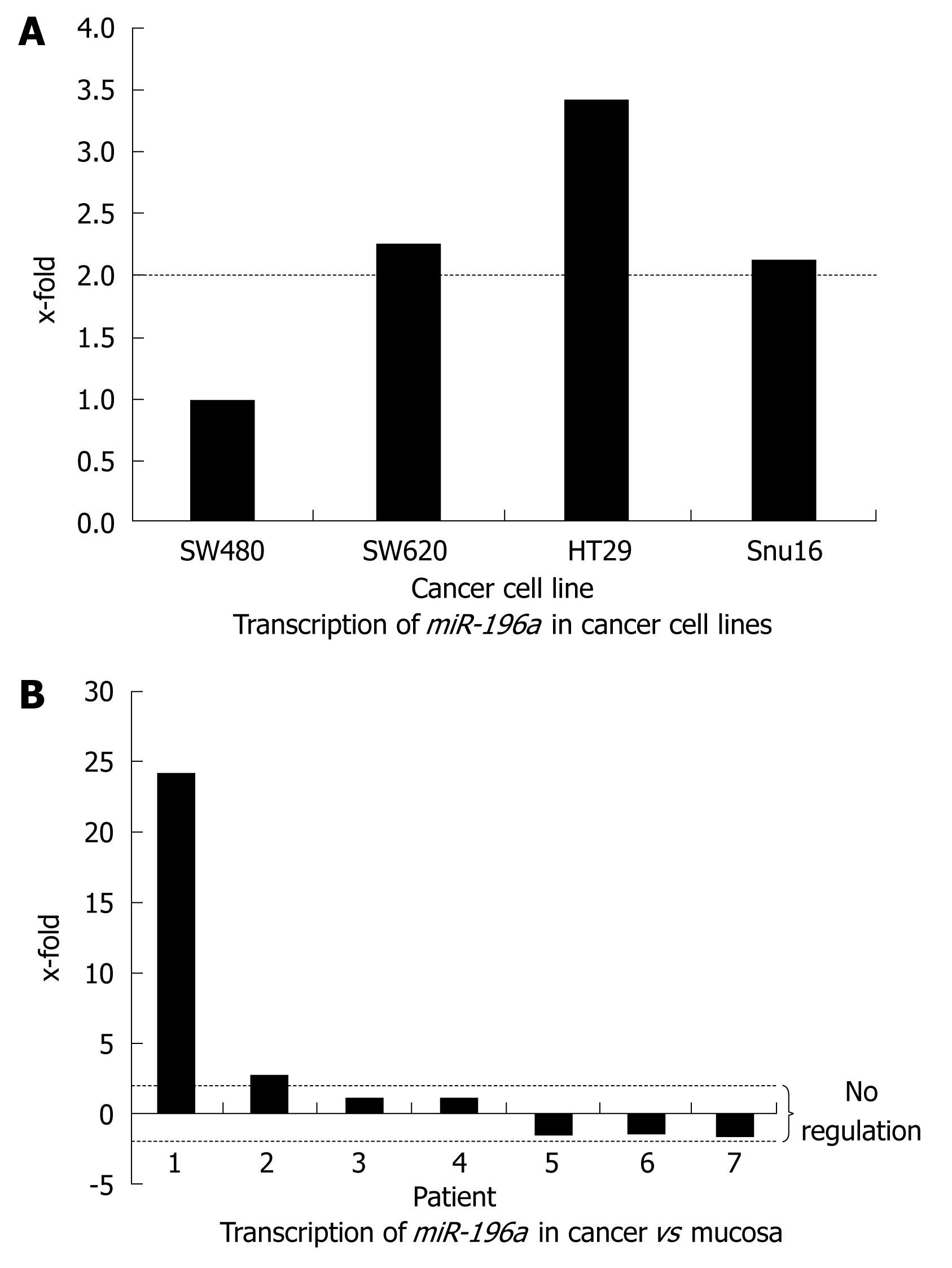
Real-time analyses of four cancer cell lines revealed U6 adjusted differences in regulation of miR-196a (Figure 1A). The SW480 cell line, which was initially isolated from a primary colon cancer, revealed the weakest transcription level. In contrast, SW620 cells, isolated from metastases of the same patient depicted a 2.25-fold up-regulation of miR-196a. HT-29, another colorectal cancer cell line revealed a 3.38-fold up-regulation of miR-196a. Similarly, SNU16 generated from metastases of a disseminated gastric cancer showed a 2.14-fold up-regulation of miR-196a.
Real-time analyses of colon cancer and matching mucosa revealed an U6 adjusted up-regulation of miR-196a in two of seven colon cancers samples analyzed (24.3- and 2.5-fold, respectively; Figure 1B). In contrast, five of seven samples did not depict any transcription differences between tumor and mucosa (1.14-, 1.04-, -1.03-, -1.08- and -1.28-fold regulation, respectively).
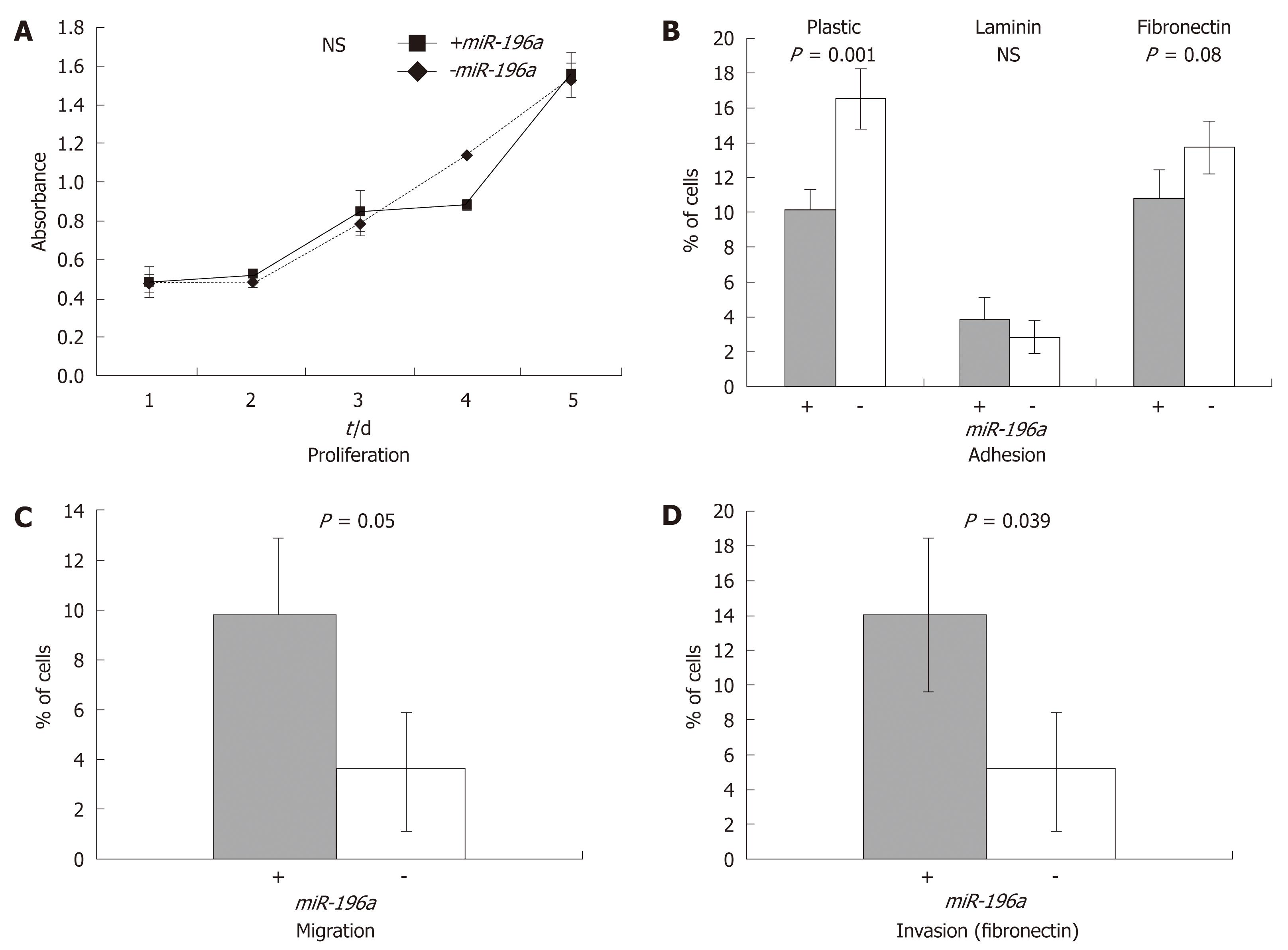
Functional analyses did not depict any significant impact of miR-196a on proliferation (Figure 2A). Absorbance analyses after 4 d of cell culture revealed the following results: +miR-196a: 1.506 ± 0.079, -miR-196a: 1.533 ± 0.131; P = 0.66; (vs NS).
Interestingly, transfection with miR-196a decreased the adhesion of cancer cells to plastic and fibronectin but not to laminin (Figure 2B). Adhesion analyses revealed following results: for plastic surface: +miR-196a: 10.2% ± 1.15%, -miR-196a: 16.6% ± 1.73%; P = 0.001. For laminin coating: +miR-196a: 3.86% ± 1.3%, -miR-196a: 2.84% ± 0.95%; P = 0.25; (vs NS) and for fibronectin coating: +miR-196a: 10.86% ± 1.64%, -miR-196a: 13.8% ± 1.56%; P = 0.08; (NS).
In addition, miR-196a transfection resulted in a significant increase of migration and invasion (Figure 2C and D): Migration: +miR-196a: 9.7% ± 3% vs -miR-196a: 3.6% ± 2.4%; P = 0.05. Invasion: +miR-196a: 12.6% ± 3% vs -miR-196a: 5.14% ± 3%; P = 0.039.
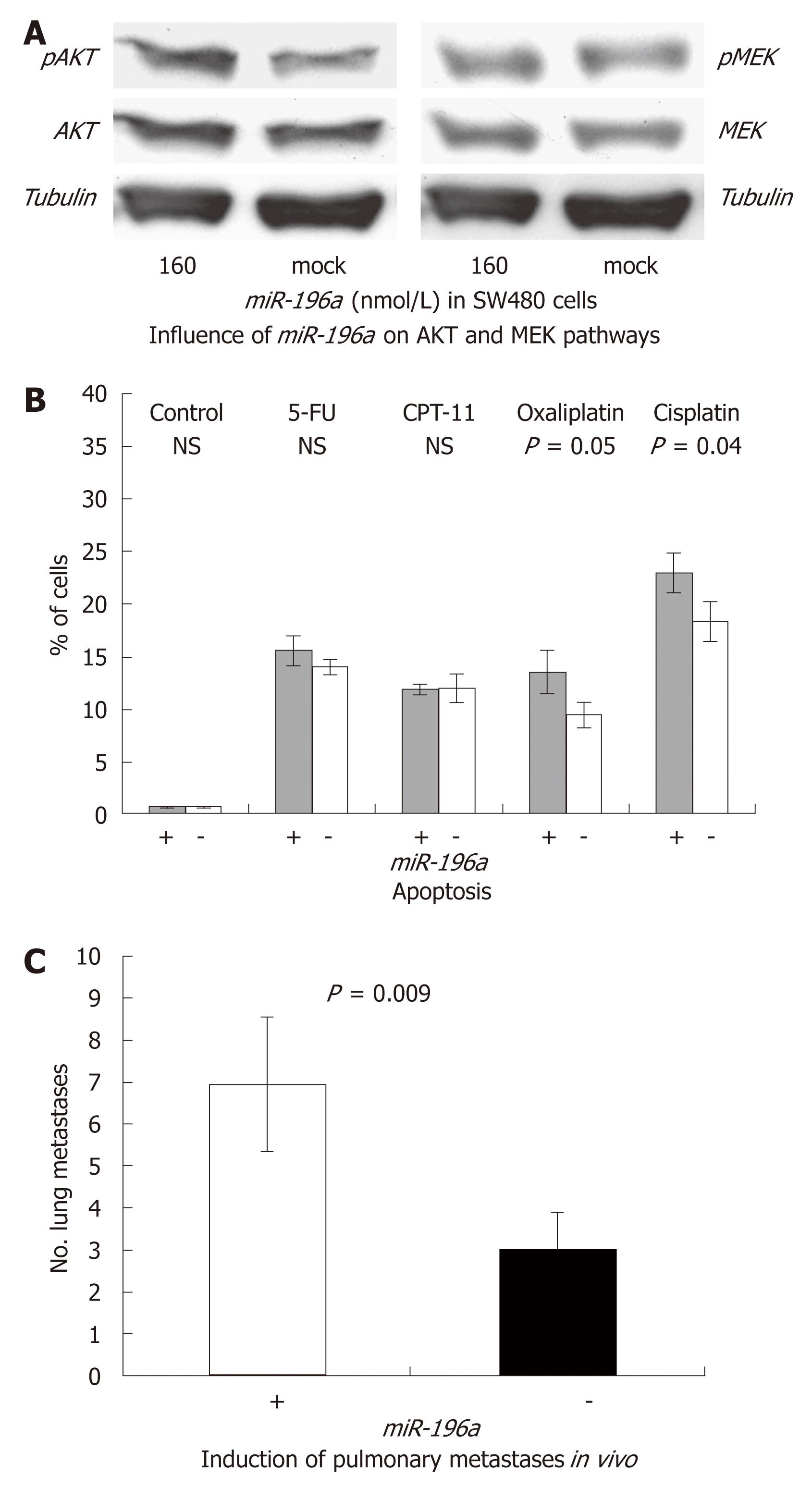
In order to analyze the relevance of miR-196a on activation of signal cascades we quantified phosphorylation of AKT and MEK (Figure 3A). Transient transfection with miR-196a resulted in an increased phosphorylation of (p)AKT but not of (p)MEK. These results imply that miR-196a increases activation of the PI3K-AKT-mTor signalling pathway.
Analyses of apoptosis did not reveal any significant impact of miR-196a (Figure 3B): +miR-196a: 0.61% ± 0.08% vs -miR-196a: 0.62% ± 0.07%, P = 0.3; (NS); nor in combination with 5-FU [+miR-196a: 15.67% ± 1.45% vs -miR-196a: 14.05% ± 0.74%, P = 0.18; (NS)] or irinotecan [+miR-196a: 11.97% ± 0.51% vs -miR-196a: 12.06% ± 1.36%, P = 0.92; (NS)]. However, miR-196a significantly increased chemosensitivity to oxaliplatin (+miR-196a: 13.56% ± 2.08% vs -miR-196a: 9.46% ± 1.19%, P = 0.05) and cisplatin (+miR-196a: 23.11% ± 1.93% vs -miR-196a: 18.42% ± 1.92%; P = 0.04). In summary, miR-196a increases chemosensitivity to platin derivates.
Transient transfection of SW480 cancer cells with miR-196a resulted in a significant increase of pulmonary metastases growth after 7 wk of incubation: +miR-196a: 7.5 ± 1.7 vs -miR-196a: 3.25 ± 0.96, P = 0.009 (Figure 3C).
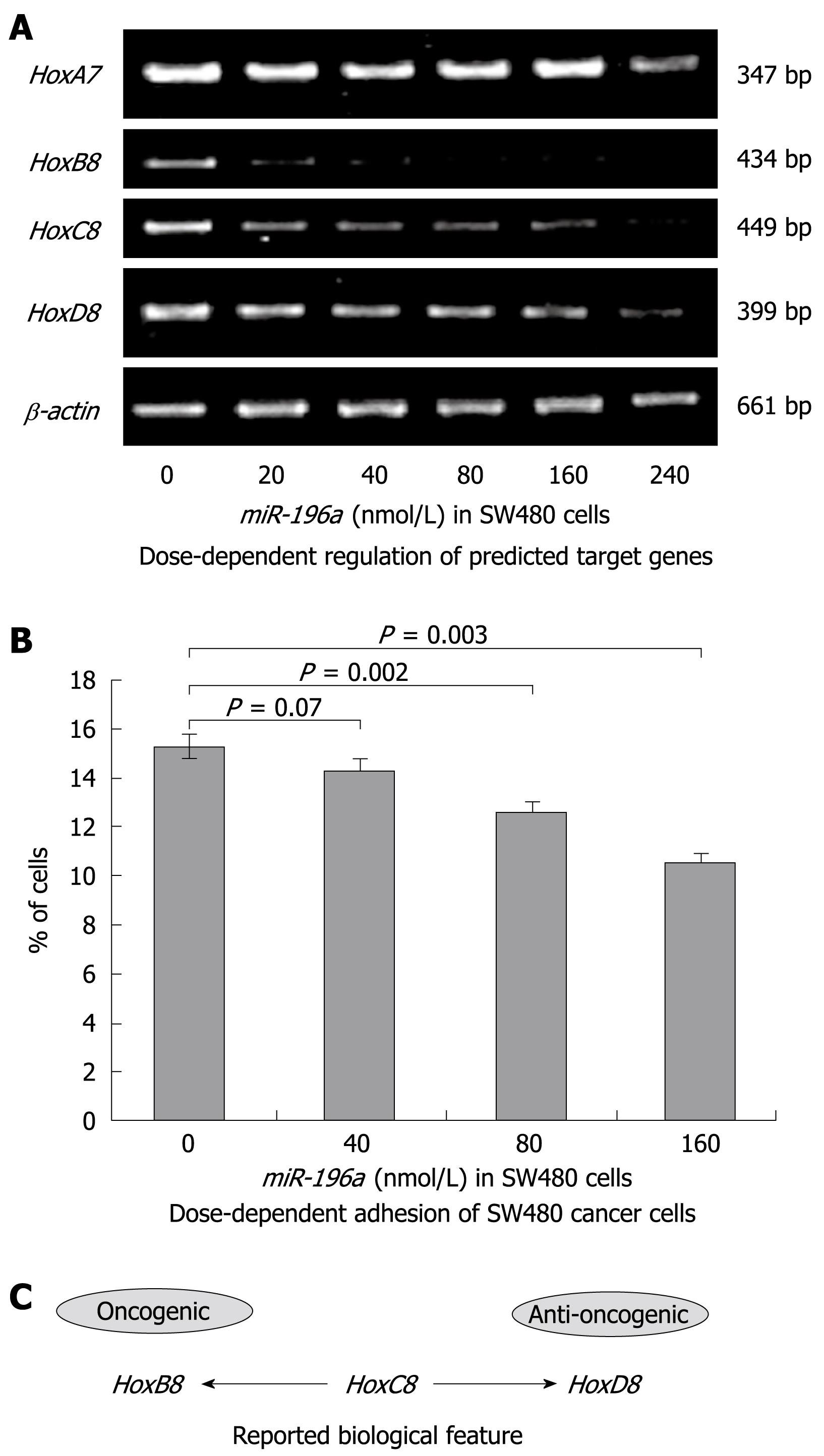
Transient transfection of SW480 cells with miR-196a verified HoxA7, HoxB8, HoxC8 and HoxD8 as miR-196a targeted genes (Figure 4A). However, significant differences in target restriction were observed. While low miR-196a concentrations (20 nmol/L) sufficiently restricted HoxB8 mRNA, higher concentrations were necessary to completely restrict HoxC8 mRNA and to restrict a significant amount of HoxD8 mRNA. However, the impact of miR-196a on HoxD8 was weaker than on HoxC8. Only the highest miR-196a concentrations (240 nmol/L) decreased mRNA levels of HoxA7. These data verify the predicted Hox genes HoxA7, HoxB8, HoxC8 and HoxD8 as human targets of miR-196a but also reveal dose-dependent differences in restriction of target genes.
Transfection with miR-196a significantly decreased the adhesion of cancer cells to plastic in a dose-dependent manner. Numbers reflect the percentage of cells that adhered to the bottom of the well: 0 nmol/L miR-196a: 15.21% ± 0.47%; 40 nmol/L miR-196a: 14.27% ± 0.46%; P = 0.07; (NS); 80 nmol/L miR-196a: 12.43% ± 0.42%; P = 0.002 and 160 nmol/L miR-196a: 10.6% ± 0.3%; P = 0.0003 (Figure 4B).
Expression patterns of miRNAs are systematically altered in colon cancer as recently described by Schetter and colleagues[25]. In particular, Schetter et al[25] reported that at least 37 miRNAs are differentially expressed in colon cancer. Of those the expression profiles of miR-20a, miR-21, miR-106a, miR-181b and miR-203 were validated. Interestingly, high miR-21 expression was associated with poor survival.
We were interested in the relevance of miR-196a transcription for human colorectal cancer progression for specific reasons. Yekta and colleagues described HoxB8 as a restriction target of miR-196a and predicted HoxA7, HoxC8 and HoxD8 as additional restriction targets in humans[22]. Hox genes are known to be master regulators of embryogenesis and oncogenesis[15]. We were able to confirm these data presented by Yekta and colleagues, as mRNA levels of those four Hox genes were reduced by miR-196a. However, dose-dependent differences in target restriction were observed. While low miR-196a concentrations resulted in a complete restriction of HoxB8 mRNA, higher concentrations of miR-196a were mandatory to completely restrict HoxC8 mRNA and to significantly decrease HoxD8 mRNA levels. In contrast, even the highest miR-196a concentrations did not result in a complete restriction of HoxA7. These data clearly reveal mRNA specific and dose-dependent target restriction. To clarify the dose-dependence of miR-196a we performed adhesion assays after transfection with different concentrations of miR-196a. These assays revealed a dose-dependent inhibition of tumor cell adhesion.
To further analyze the impact of miR-196a on tumor cells, we then performed functional assays and found that high miR-196a concentrations increased migration and invasion of cancer cells in trans-well assays and inhibited adhesion to different surfaces and matrix proteins. Chemosensitivity assays with standard chemotherapeutics revealed that miR-196a does not sensitise against 5-FU nor irinotecan, but does sensitize against the platin derivatives oxaliplatin and cisplatin. However, miR-196a did not impact on proliferation or apoptosis of colon cancer cells.
Analyzing signaling cascades that are often altered in human cancer, we observed that induction of the pro-migratory phenotype is most likely linked to activation of the PI3K-AKT-mTor pathway, as miR-196a increased the level of pAKT. In contrast, no change in the pMEK/MEK ratio was observed. Our data are consistent with earlier reports showing that overexpressed miRNAs can act as oncogenes. A well known example is miR-17-92, which is significantly increased in small-cell lung cancer and correlates with a poor prognosis[20]. Interestingly, the known targets of the miR-17-92 include the two tumor suppressor genes PTEN and RB2[21]. As a consequence, restriction of PTEN unleashes the PI3K-AKT-mTor pathway as also observed for miR-196a. However, the exact mode of action of miR-196a has still to be analyzed.
Quantitative real-time PCR of miR-196a in matching colon cancer and colon mucosa samples showed an up-regulation in 28% of samples. In contrast, all other cancer samples revealed no regulation at all. Most interestingly, the metastatic cancer cell lines SW620 and HT29 showed a significant up-regulation of miR-196a in contrast to SW480 cells isolated from a primary colon cancer. Therefore, miR-196a is up-regulated in a subset of colorectal cancers and might exert an oncogenic function, when transcribed at a high level. Matching these observations, Croce and colleagues recently found that 75% of pancreatic cancers expressed miR-196a at a high level, predicting poor patient survival (14.3 mo vs 26.5 mo) when investigating the miRNA transcription pattern in pancreatic adenocarcinoma with large scale miRNA chips[24]. Therefore, similar mechanisms seem possible for pancreatic and colorectal cancer.
To verify the oncogenic potential of high miR-196a concentrations, we further analyzed the impact of miR-196a in an in vivo lung metastases xenograft bio-system. After transient transfection of cells with high concentrations of miR-196a prior to tail-vein injection, mice developed significantly more pulmonary metastases within 7 wk as compared to mock-transfected cells.
In summary, we observed an oncogenic effect of high miR-196a concentrations. However, several data imply that miR-196a might function as a double-edged sword with opposing effects at different concentration for following reasons. (1) miR-196a is transcribed in colon mucosa at low levels, implying a role for the epithelial phenotype. (2) A hypothesized suppressive effect of low miR-196a transcription levels on tumor dissemination might be exerted through a dose-dependent restriction of miR-196a target genes HoxB8, HoxC8 and HoxD8. Up-regulation of HoxC8 and HoxB8 in colorectal cancer was reported as early as 1997, however the relevance of those genes for carcinogenesis had not been analyzed[26]. A relevant leukemogenic property of HoxB8 mediated through inhibition of differentiation has been described for acute myeloid leukemia[2728]. These data are intriguing, as low concentrations of miR-196a completely restrict HoxB8, thus erasing the pro-oncogenic and leukemogenic effects of HoxB8. (3) Only very limited data concerning the relevance of HoxD8 is available, indicating that HoxD8 are up-regulated after chemical induced re-differentiation of neuroblastoma cells[29]. However, this observation is of particular interest, as high miR-196a concentrations are needed to significantly reduce HoxD8 mRNA levels, which might result in an inhibition of differentiation, thus promoting oncogenic features as observed in our analyses. (4) The data concerning the relevance of HoxC8 is unclear. Both pro- and anti-oncogenic influences have been discussed. In particular, HoxC8 was reported to be a retinoic acid induced gene, rescuing APC mutants in zebrafish[30]. In contrast, studies on prostate cancer have reported a correlation with aberrant HoxC8 expression and a malignant phenotype[3132]. As Hox genes are master transcription factors, they might exert different functions at variable expression levels. However, the observation of Croce and colleagues that miR-196a predicts poor survival in pancreatic cancer might rather correlate with inhibition of HoxD8 than HoxB8 expression, as HoxD8 has a suppressive and HoxB8 a progressive character in the literature[24]. Further studies analyzing the clinical and biological impact of miR-196a, as well as additional large scale analyses of restriction targets, are warranted.
MicroRNAs (miRNAs) are small RNA molecules regulating gene expression in vertebrae and non-vertebrae. In humans, more than 50% of all known miRNA genes are located in cancer-associated regions, indicating that miRNAs might play an important role in oncogenesis. Some miRNAs are known to function as oncogenes, while others act as tumor suppressors inhibiting tumor growth.
Hox proteins are major transcription factors that play a crucial role during embryogenesis, organogenesis and oncogenesis. The miRNA miR-196a depicts complementarity to the mRNA of HoxB8, HoxC8 and HoxD8. Therefore, the relevance of miR-196a for human tumorigenesis has been discussed.
High levels of miR-196a activated oncogenic pathways inside the human tumor cells and induced tumor cell detachment, migration and invasion. In addition, miR-196a promoted growth of lung metastases in mice. However, miR-196a also increased the chemosensitivity towards platin derivatives such as cisplatin and oxaliplatin.
High levels of miR-196a might predict response of cisplatin- or oxaliplatin-containing chemotherapies. In future, suppression of miR-196a by anti-miR technologies might inhibit tumor progression and dissemination.
miRNAs are endogenous small RNA molecules of 20-25 nucleotides length, regulating gene expression by inhibiting transcription, inducing direct cleavage of the targeted mRNAs or blocking translation through their complementarity versus targeted mRNAs at 3′ untranslated regions.
This is a very interesting study which contributes to our understanding of colorectal cancer, its development and prognosis. The paper is well written.
| 1. | Weir HK, Thun MJ, Hankey BF, Ries LA, Howe HL, Wingo PA, Jemal A, Ward E, Anderson RN, Edwards BK. Annual report to the nation on the status of cancer, 1975-2000, featuring the uses of surveillance data for cancer prevention and control. J Natl Cancer Inst. 2003;95:1276-1299. |
| 2. | Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7-33. |
| 3. | August DA, Ottow RT, Sugarbaker PH. Clinical perspective of human colorectal cancer metastasis. Cancer Metastasis Rev. 1984;3:303-324. |
| 5. | Cho KR, Vogelstein B. Genetic alterations in the adenoma--carcinoma sequence. Cancer. 1992;70:1727-1731. |
| 6. | Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138-141. |
| 7. | Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433-9438. |
| 10. | Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336-338. |
| 11. | de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3' UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49-58. |
| 12. | Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133-1146. |
| 13. | Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363-364. |
| 14. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. |
| 15. | Wynter CV. The dialectics of cancer: A theory of the initiation and development of cancer through errors in RNAi. Med Hypotheses. 2006;66:612-635. |
| 16. | Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753-3756. |
| 17. | Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903-906. |
| 18. | Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293-300. |
| 19. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. |
| 20. | Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628-9632. |
| 21. | Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-798. |
| 22. | Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-596. |
| 23. | Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671-674. |
| 24. | Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901-1908. |
| 25. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. |
| 26. | Vider BZ, Zimber A, Chastre E, Gespach C, Halperin M, Mashiah P, Yaniv A, Gazit A. Deregulated expression of homeobox-containing genes, HOXB6, B8, C8, C9, and Cdx-1, in human colon cancer cell lines. Biochem Biophys Res Commun. 2000;272:513-518. |
| 27. | Knoepfler PS, Sykes DB, Pasillas M, Kamps MP. HoxB8 requires its Pbx-interaction motif to block differentiation of primary myeloid progenitors and of most cell line models of myeloid differentiation. Oncogene. 2001;20:5440-5448. |
| 28. | Perkins AC, Cory S. Conditional immortalization of mouse myelomonocytic, megakaryocytic and mast cell progenitors by the Hox-2.4 homeobox gene. EMBO J. 1993;12:3835-3846. |
| 29. | Manohar CF, Salwen HR, Furtado MR, Cohn SL. Up-regulation of HOXC6, HOXD1, and HOXD8 homeobox gene expression in human neuroblastoma cells following chemical induction of differentiation. Tumour Biol. 1996;17:34-47. |
| 30. | Nadauld LD, Sandoval IT, Chidester S, Yost HJ, Jones DA. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J Biol Chem. 2004;279:51581-51589. |
| 31. | Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162-169. |
| 32. | Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, Schirripa O, Lucia MS, Nordeen SK. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879-5888. |