Published online Apr 28, 2009. doi: 10.3748/wjg.15.2020
Revised: February 23, 2009
Accepted: March 2, 2009
Published online: April 28, 2009
AIM: To explore the relationship between DNA methyltransferase 1 (DNMT1) and hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) and its biological significance in primary HCC.
METHODS: We carried out an immunohistochemical examination of DNMT1 in both HCC and paired non-neoplastic liver tissues from Chinese subjects. DNMT1 mRNA was further examined in HCC cell lines by real-time PCR. We inhibited DNMT1 using siRNA and detected the effect of depletion of DNMT1 on cell proliferation ability and cell apoptosis in the HCC cell line SMMC-7721.
RESULTS: DNMT1 protein expression was increased in HCCs compared to histologically normal non-neoplastic liver tissues and the incidence of DNMT1 immunoreactivity in HCCs correlated significantly with poor tumor differentiation (P = 0.014). There were more cases with DNMT1 overexpression in HCC with HBV (42.85%) than in HCC without HBV (28.57%). However, no significant difference in DNMT1 expression was found in HBV-positive and HBV-negative cases in the Chinese HCC group. There was a trend that DNMT1 RNA expression increased more in HCC cell lines than in pericarcinoma cell lines and normal liver cell lines. In addition, we inhibited DNMT1 using siRNA in the SMMC-7721 HCC cell line and found depletion of DNMT1 suppressed cells growth independent of expression of proliferating cell nuclear antigen (PCNA), even in HCC cell lines where DNMT1 was stably decreased.
CONCLUSION: The findings implied that DNMT1 plays a key role in HBV-related hepatocellular tumorigenesis. Depletion of DNMT1 mediates growth suppression in SMMC-7721 cells.
- Citation: Fan H, Zhao ZJ, Cheng J, Su XW, Wu QX, Shan YF. Overexpression of DNA methyltransferase 1 and its biological significance in primary hepatocellular carcinoma. World J Gastroenterol 2009; 15(16): 2020-2026
- URL: https://www.wjgnet.com/1007-9327/full/v15/i16/2020.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.2020
DNA methylation plays an important role in transcriptional regulation, chromatin remodeling and genomic stability[1–3]. It is catalyzed in mammalian cells by a family of highly related DNA methyltransferases (DNMTs) that use S-adenosylmethionine as the methyl donor[4]. Alteration of DNA methylation is one of the most consistent epigenetic changes in human cancers and is involved even in the early and precancerous stages of human carcinogenesis[56]. Overexpression of DNMT1 has been detected in several human cancers[7–11] and showed a relatively significant correlation with tumorigenesis, but not in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). HCC is a devastating disease with a very poor prognosis, and is the fourth or fifth largest cause of cancer-related death worldwide[1213]. More than 85% of Chinese HCC patients have HBV infection history. Data on hepatocarcinogenesis in other countries showed that DNMT1 mRNA and protein expression were significantly higher in HCCs[1415]. Therefore, DNMT1 may play an important role during hepatocarcinogenesis. However, the relationship between HBV infection and DNMT1 in HCC has still to be elucidated, especially in Chinese subjects. In the present study, we examined the expression of DNMT1 both in HBV-related HCC cases and cell lines from Chinese subjects. DNMT1 expression was significantly increased both in tumor tissues and HCC cell lines compared with corresponding controls. To further investigate and evaluate the possibility of DNMT1 knockdown strategies for cancer therapy, we inhibited DNMT1 using siRNA and examined the cell growth and cell proliferation ability in the HCC cell line SMMC-7721.
We selected 42 cases of surgically resected livers from the surgical pathology files of the Department of Pathology at The Qidong County Hospital, China. The patients’ clinicopathologic features are shown in Table 1. All the HCC samples were diagnosed by a single pathologist and appropriate consent was obtained. Paired samples of primary HCC and matched non-cancerous normal tissues were obtained from each patient. The pathological classification of HCC tissues was carried out and the stage of each HCC was determined according to criteria. Histological examination of noncancerous liver tissues from HCC patients revealed no remarkable findings. Thirty-five cases were associated with HBV infection, and 7 cases were negative for HBV.
| Patients and tissue specimens | n |
| Sex | |
| Male | 36 |
| Female | 6 |
| Viral status | |
| HBs-Ag positive | 35 |
| HBs-Ag negative | 7 |
| Histology of noncancerous liver tissues | |
| Histologically normal | 18 |
| Liver cirrhosis | 24 |
| Tumor differentiation | |
| Well differentiated | 0 |
| Moderately differentiated | 22 |
| Poorly differentiated | 20 |
| Total | 42 |
Nine human cell lines were cultured for the study. Human hepatocellular carcinoma cell lines (BEL-7402, BEL-7404, BEL-7405, QGY-7703, QGY-7701, SMMC-7721), an immortalized human hepatocellular normal cell line (HL-7702) or a pericarcinoma (QSG-7701) cell line were cultured in RPMI-1640 (Life Technologies, Inc., Rockville, MD) containing 10% new born bovine serum in 5% CO2 incubation at 37°C.
siRNAs targeting DNMT1 were designed and prepared as described previously[16]. The siRNA sequences against DNMT1 were designed as sense and antisense oligonucleotides corresponding to nucleotide position 2620-2638 of human DNMT1 (GenBank accession No. NM001379.1). The siRNA sequence and scrabble sequence were sub-cloned into the pSUPER-EGFP vector (gift from Dr. Dianqing Wu), which was identified by HindIII (TAKARA) and BglII (TAKARA), to be the DNMT1 siRNA construct named pMT1, with sMT1 as a control.
The human HCC cell line SMMC-7721 (No. TCHu13 Cell Bank Shanghai, China) was cultured in RPMI-1640 containing 10% new born bovine serum in 5% CO2 incubation at 37°C. Cells were transfected with 1.5 &mgr;g of DNMT1 siRNA (pMT1) construct or sMT1 using transfectamineTM 2000 reagent (Invitrogen), and selected with 0.4 mg/mL genetincin (Life Technologies). SMMC-7721 cells were transfected with pMT1 and named 7721-MT1 cell lines or transfected with sMT1 and named 7721-sMT1 as a control.
Four micrometre thick sections of formalin-fixed, paraffin embedded tissue specimens from all 42 patients were deparaffinized and dehydrated. For antigen retrieval, the sections were heated for 10 min at 120°C in an autoclave, and nonspecific reactions were blocked with 5% normal horse serum. All sections were incubated with specific primary antibodies that recognized DNMT1 (goat polyclonal antibody, dilution 1:500; Santa Cruz Biotechnology, Santa Cruz, CA). We previously confirmed the specificity of the goat anti-human DNMT1 polyclonal antibody by Western blotting analysis: an immunoreactive band of 193.5 kDa, corresponding to the molecular mass of DNMT1, was detected in human cancer cells, but no nonspecific bands were detected with this antibody[16]. All primary antibody incubations were conducted at 4°C overnight and were followed by incubation with biotinylated secondary antibodies (anti-goat IgG, anti-mouse IgG, dilution 1:200; QIAGEN Laboratories) at room temperature for 30 min. The sections were then treated with Vectastain Elite ABC reagent (Vector Laboratories). All sections were counterstained with hematoxylin. For negative control preparations, the primary antibody was omitted from the reaction sequence.
Total RNA was purified from a normal liver cell line, a cell line established from pericarcinoma tissue and HCC cell lines with TRIzol (Invitrogen). The first-strand cDNA was synthesized from 2 &mgr;g total RNA using Oligo (dT) 18 primer and SuperScript II reverse transcription kit (Life Technologies). A PCR reaction was performed in a 50 &mgr;L volume with 5U polymerase (TAKARA) and cDNA samples equivalent to 1 ng of RNA. SYBR green with 20 000 dilutions was included in each reaction for relative quantification in the ABI 7300 sequence detection system (Applied Biosystems). To normalize the input load of cDNA among samples, β-actin was quantified and used as an endogenous standard. The relative level of expression of each DNMT1 among different cell lines was then calculated accordingly (ABI PRISM 7300 Sequence Detection System, USA). The primers used for PCR were as follows: DNMT1: sense primer, 5'-CCGAGTTGGTGATGGTGTGATC-3′; antisense primer, 5′-AGGTTGATGTCTGCGTGGTAG C-3′. β-actin: 5′-AAAGACCTGTACGCCAACAC-3′; antisense primer, 5′-GTCATACTCCTGCTTGCTGAT-3′. PDCD4: sense primer 5′-TGGATGAAAGGGCATTTGAGA-3′; antisense primer, 5′-AGCCTTCCCCTCCAATGCTA-3′.
Cells were grown and harvested at 80%-85% confluency, and cellular proteins were extracted with lysis buffer containing 0.5% NP-40, 150 mmol/L NaCl, and 1 mmol/L EDTA in 50 mmol/L Tris-HCl at pH 7.5, supplemented with a protease inhibitor cocktail (Sigma Chemical Co., St. Louis, MO). The protein concentration of each extract was quantified by BCA assay (Pierce, Rockford, IL). Two to forty microliter of total protein was electrophoresed on 7%-15% SDS-polyacrylamide gel and transferred to polyvinylidene fluoride membranes (PVDF, Amershan) electrophoretically. After blocking with 5% nonfat dry milk and 0.1% Tween 20 in Tris-buffered saline, membranes were incubated with goat anti-DNMT1 (Santa Cruz Biotechnology), proliferating cell nuclear antigen (PCNA) (mouse monoclonal antibody, dilution 1:200, Lexington, KY) or mouse monoclonal anti-actin (Sigma) antibodies. The membranes were then developed with peroxidase-labeled antibodies (Amersham Pharmacia, Piscataway, NJ) by Super Signal chemiluminescence substrate (Pierce, Rockford, IL). Actin protein levels were used as a control for equal protein loading.
Cell proliferation of transiently transfected SMMC-7721 cells was measured by trypan blue dye cell count assay. Cells were cultured in triplicate in 12-well plates at a concentration of 3 × 104 cells per well. Cells were collected at 1, 3, 5 and 7 d and exposed to trypan blue (Sigma), and nonviable cells took up the dye. Both viable (unstained) and nonviable (stained) cells were counted, and the relative survival rate (%) of each siRNA treatment was then calculated.
Stably transfected SMMC 7721 cells were washed, resuspended in staining buffer, and examined by ApoAlert Annexin V Apoptosis kit (BD Biosciences) and PI according to the manufacturer’s instructions. Stained cells were analyzed by FACS (FACScalibur, BD Biosciences).
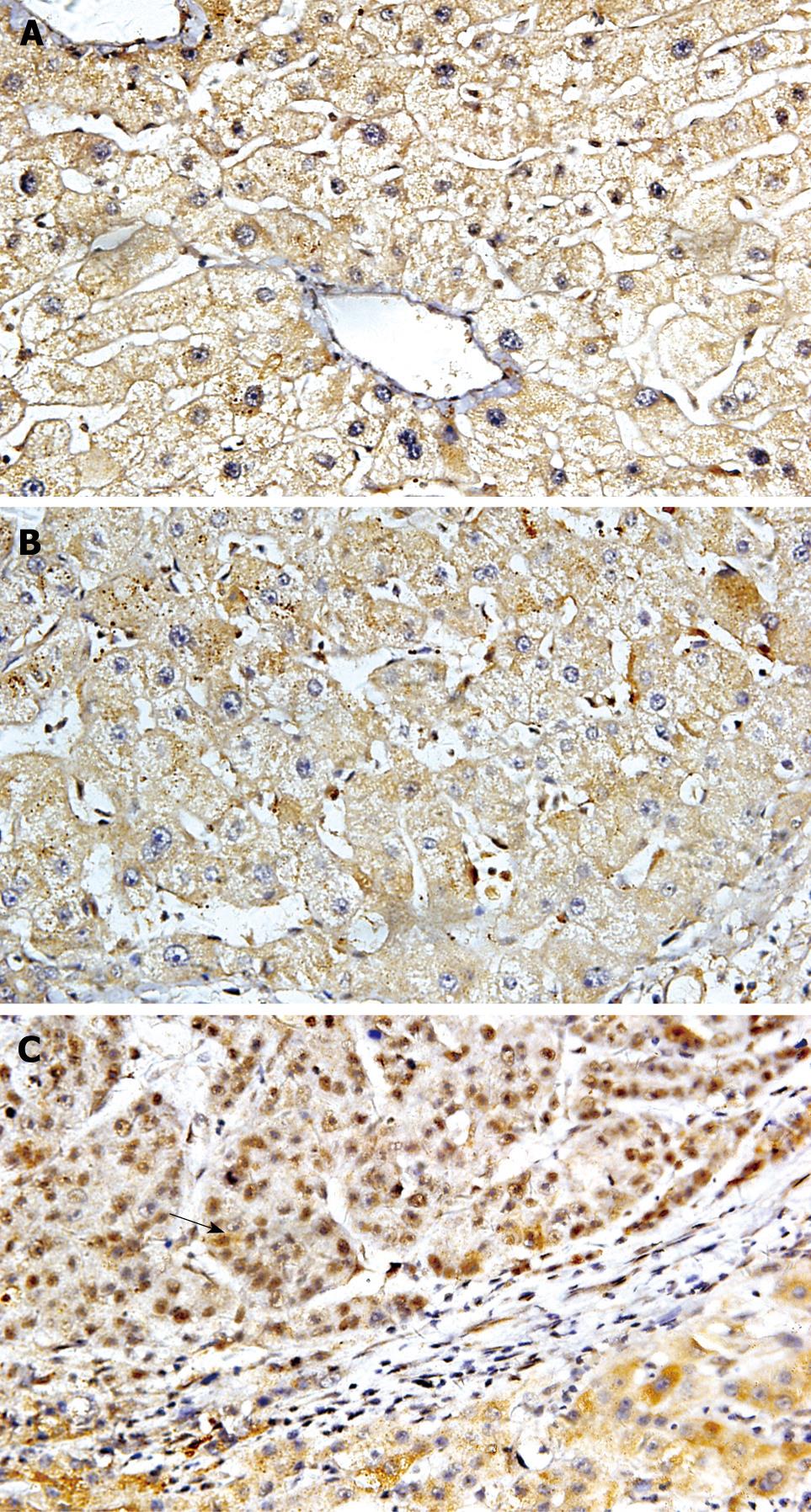
Immunoreactivity for DNMT1 was detected in the nuclei and cytoplasm, but not in the cell membranes, of cancer cells (Figure 1). To discriminate definitely positive cases from cases with a leaky background level signal, if more than 30% of the cells in a tissue sample exhibited nuclear and cytoplasm staining the sample was considered to show positive immunoreactivity. Nuclear and cytoplasmic DNMT1 immunoreactivity was detected and overexpressed in 40.48% of patients. The incidence of DNMT1 immunoreactivity in carcinoma correlated significantly with poor tumor differentiation (Table 2, P = 0.014, χ2 test). The incidence of DNMT1 immunoreactivity was significantly higher in HCCs than in pericancerous liver tissues. DNMT1 protein overexpression was not significantly associated with other parameters relating to cancer aggressiveness, such as the depth of invasion, vascular involvement, or lymph node metastasis. In HBV-related HCC cases, 70% had different degrees of hepatocirrhosis. To investigate whether there was an HBV-induced increase in DNMT1 expression in HCC, we evaluated the relationship between HBV infection and up-regulated DNMT1. There was higher expression of DNMT1 in HCC with HBV than in HCC without HBV. However, there was no significant correlation between the incidence of DNMT1 immunoreactivity in HCCs and the patients’ viral status (HBs-Ag-positive, HBs-Ag-negative) by statistical analysis.
| DNMT1 expression (P/OR: up-regulated/normal + down-regulated) | ||||||
| Subtotal | Up-regulated | Normal | Down-regulated | P | OR (95% CI) | |
| Total or subtotal | 42 (100.00) | 17 (40.48) | 13 (30.95) | 12 (28.57) | ||
| Age (yr) | 0.54 | 1.49 (0.41-5.35) | ||||
| > 50 | 15 (35.71) | 7 (46.67) | 4 (26.67) | 4 (26.67) | ||
| < 50 | 27 (64.29) | 10 (37.04) | 9 (33.33) | 8 (29.63) | ||
| Size of tumor (cm) | 0.23 | 0.45 (0.12-1.67) | ||||
| > 5 | 17 (40.48) | 5 (29.41) | 6 (35.29) | 6 (35.29) | ||
| < 5 | 25 (59.52) | 12 (48.00) | 7 (28.00) | 6 (24.00) | ||
| Histological differentiation | 0.014 | 0.20 (0.05-0.75) | ||||
| Moderate | 22 (52.38 ) | 5 (22.72) | 9 (40.91) | 8 (36.37) | ||
| Poor | 20 (47.62) | 12 (60.00) | 4 (20.00) | 4 (20.00) | ||
| HBsAg | 0.169 | 4.5 (0.829-24.4) | ||||
| Positive | 35 (83.33) | 15 (42.86) | 12 (34.28) | 8 (22.86) | ||
| Negative | 7 (16.67) | 2 (28.57) | 1 (14.29) | 4 (57.14) | ||
| Hepatocirrhosis | 0.083 | 3.57 (0.81-15.71) | ||||
| With | 24 (57.14) | 10 (41.67) | 10 (41.67) | 4 (16.67) | ||
| Without | 18 (42.86) | 3 (16.67) | 7 (38.89) | 8 (44.44) | ||
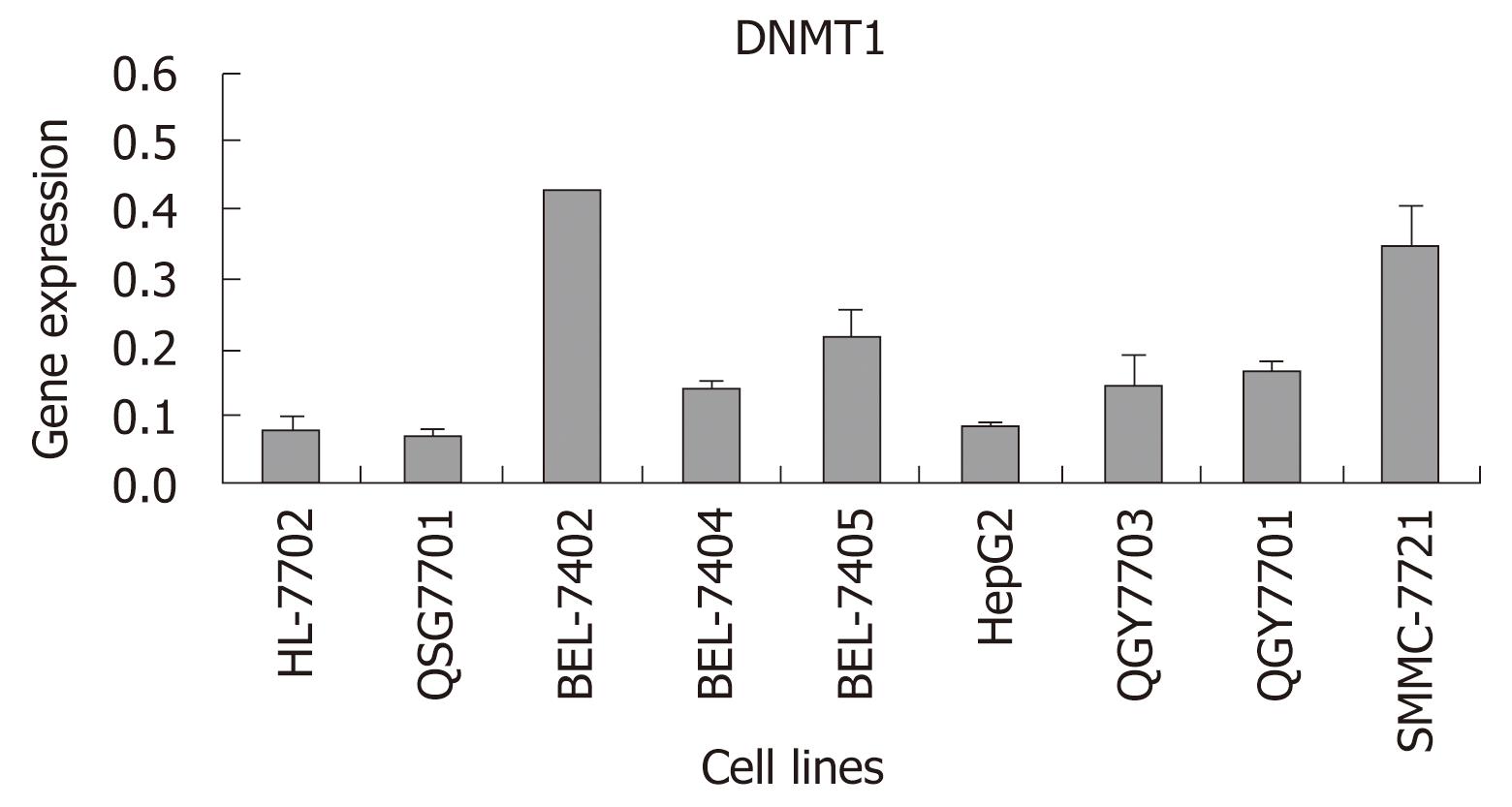
In order to determine whether the expression of DNMT1 was different between HCC cell lines and normal hepatocellular cell lines which were cultured under the same conditions, real-time RT-PCR was carried out in ABI 7300. DNMT1 was increased more in most HCC cell lines, especially in BEL-7405, BEL-7402 and SMMC-7721, than in a normal cell line and a pericarcinoma cell line. There were a trend that DNMT1 increased more in HCC cell lines than in a normal liver cell line and a pericarcinoma cell line. In contrast to the low expression level of DNMT1 in the pericarcinoma cell line, the levels of DNMT1 in most HCC cell lines were increased more than 2-fold (Figure 2).
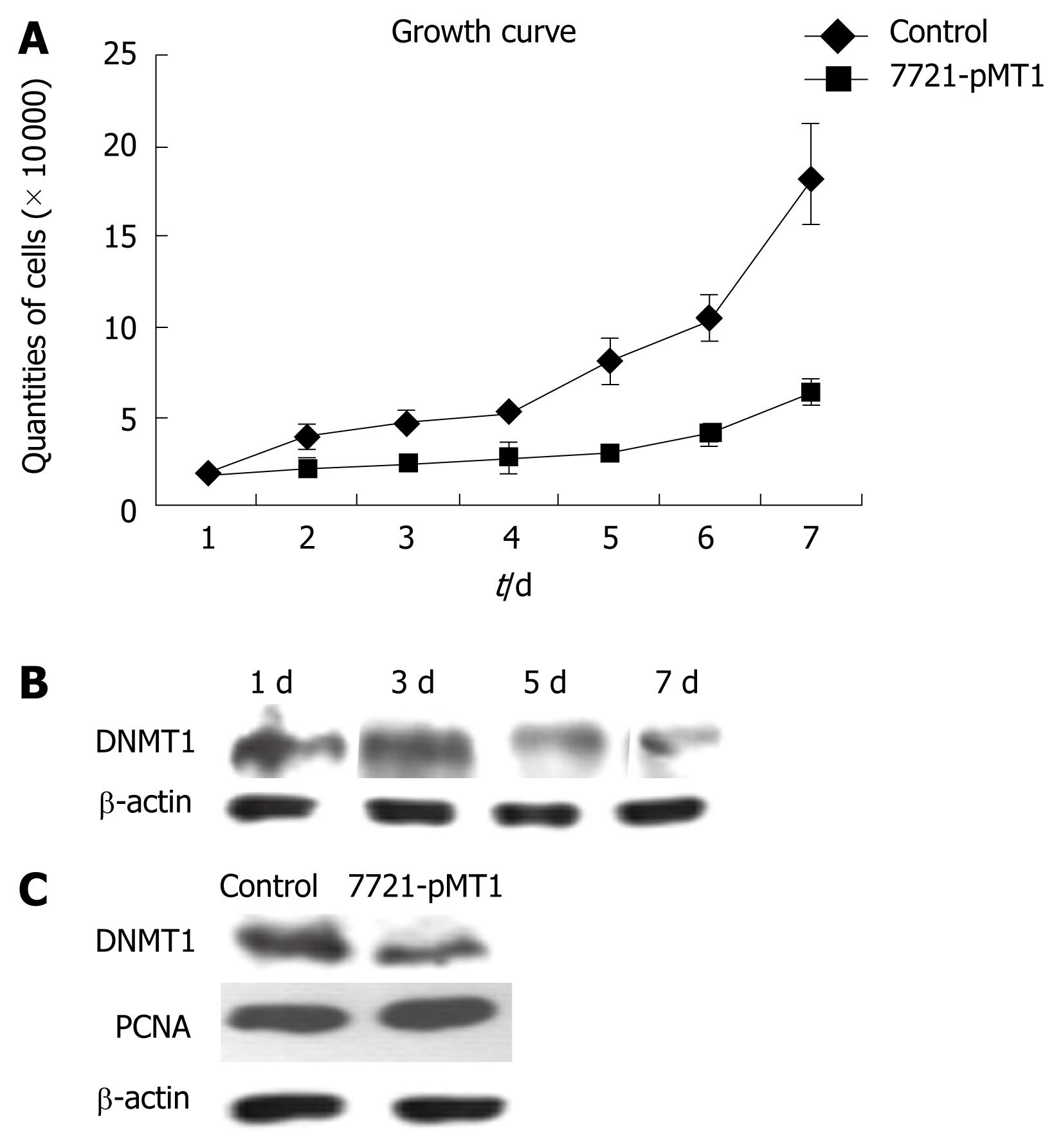
We analyzed the proliferation ability of SMMC-7721 cells transfected with DNMT1 siRNA constructs and its control by FCM and trypan blue assay. The resulting RNA interference had a negative effect on SMMC-7721 growth (Figure 3). This growth suppression was noticeable two days after transfection, and the effect was more prominent in DNMT1 knockdown cells than at seven days after transfection (Figure 3A). For the determination of DNMT1 protein expression in construct pMT1 transiently transfected SMMC 7721 cells, β-actin was employed for adjustment of DNMT1 expression data to protein content. PCNA, an established cell proliferation marker and the DNA replication factor to which DNMT1 binds, was employed to evaluate cell proliferation. Comparison with PCNA expression levels revealed that the changes in DNMT1 expression were independent of the cell proliferation status (Figure 3B). The data showed that DNMT1 levels started to decrease on day 3 and was maintained until seven days after transfection with cell growth inhibition. However, there were no significant changes in PCNA expression, even in a stably decreased DNMT1 HCC cell line (Figure 3C).
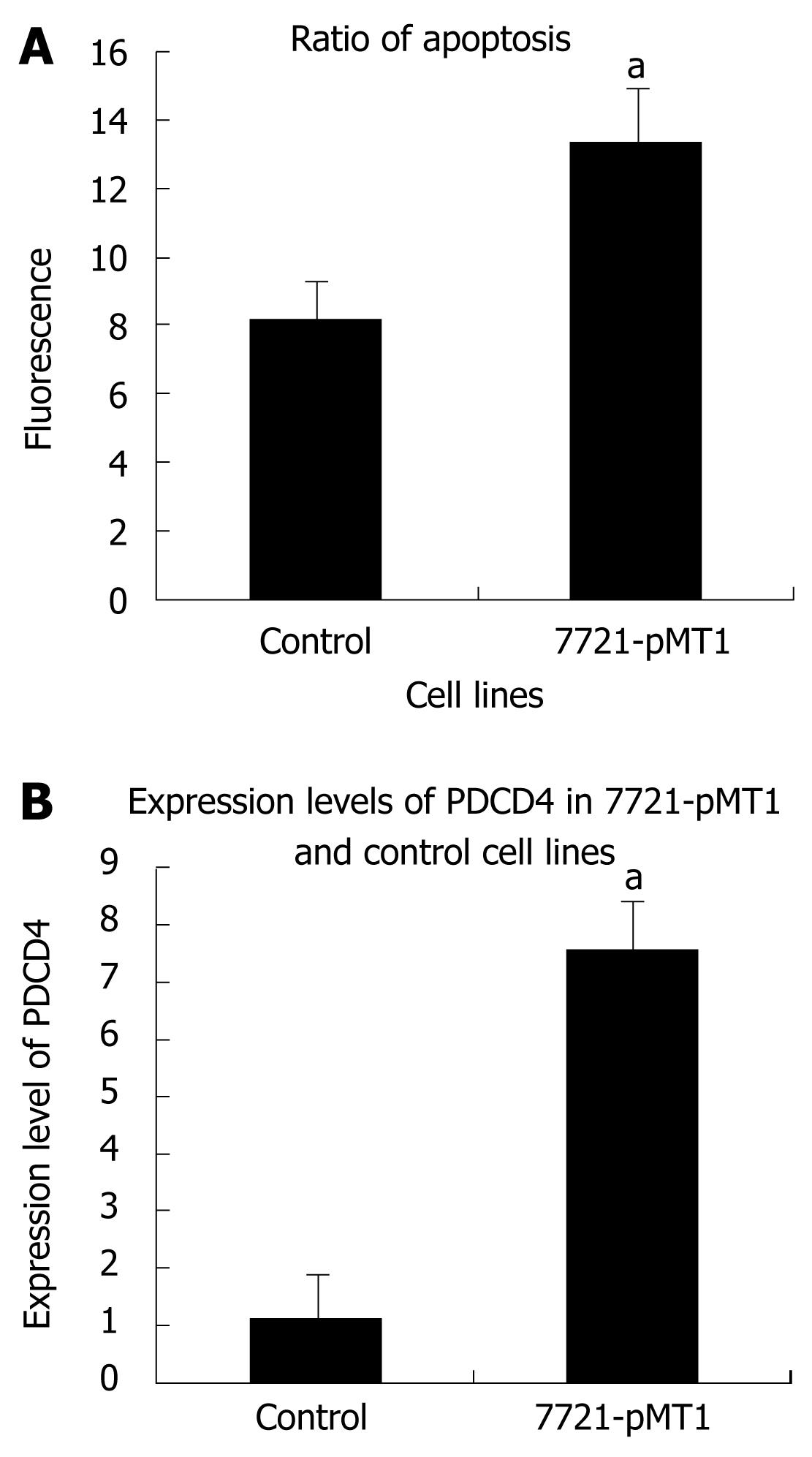
Cell apoptosis detection was performed by flow cytometric analysis as described above. The data from the present study indicated that DNMT1 knockdown induced cell apoptosis. The apoptotic rate increased from 8.78% ± 0.44% to 14.24% ± 0.12%. These results indicated that DNMT1 knockdown may induce HCC cell apoptosis (Figure 4). The apoptosis gene PDCD4 was induced by DNMT1 siRNA in SMMC-7721.
HCC is well known to develop through the stages of dysplasia and early HCC in a background of chronic liver diseases, including chronic hepatitis and liver cirrhosis. Increased DNMT1 mRNA expression has been reported in a number of human cancers[17–19]. DNMT1 may play an important role during hepatocarcinogenesis even at the precancerous stage[920]. Although DNMT1 expression and its role were evaluated in different populations, information from Chinese subjects was still unclear. HCC is one of the most lethal and prevalent cancers in China, where HBV is one of the main attributable risk factors[421]. To study the significance of aberrant DNMT1 expression during hepatocarcinogenesis in a Chinese population, we used an immunohistochemical technique to directly evaluate DNMT1 protein expression in noncancerous liver tissues and HCCs, and real-time PCR to evaluate DNMT1 expression in HCC cell lines and a normal liver cell line.
After all tumors were analyzed, HBV infection was present in 83.3% of the samples, indicating that our study cohort is representative of the subset of HCC arising from HBV infection, which is the most considerable risk factor for HCC in studied cases. In the present study, more than 80% of nonneoplastic liver tissues with either chronic hepatitis or liver cirrhosis showed variable nuclear and cytoplasm immunopositivity for DNMT1. DNMT1 immunoreactivity was certainly detected in HCCs, as it was in other cancers[22–24]. DNMT1 overexpression was detected in 40.48% of the patients and the incidence of DNMT1 immunoreactivity was significantly higher in HCC than in pericancerous liver tissues. Specifically, the incidence of DNMT1 immunoreactivity in HCCs correlated significantly with poor tumor differentiation. The above evidence indicates that increased DNMT1 protein expression may play a role in the malignant progression of HCCs in Chinese subjects. Alternatively, in virus-associated cancers, viral proteins have been shown to disturb the host DNA methylation system by up-regulating DNMT activities, thereby increasing tumor susceptibility[25]. Park et al[26] showed that HBx promoted specific regional hypermethylation of tumor suppressor genes and genome-wide hypomethylation by transcriptional regulation of DNMTs. HBx expression elevates overall intracellular DNMT activity by inducing DNMT1 and DNMT3A. HBx promotes epigenetic abnormalities by modulating the expression of DNMTs immediately after HBV infection, thus epigenetically accelerating hepatocarcinogenesis. In this study, we evaluated DNMT1 expression in HBV-positive cases and HBV-negative cases. No significant relationship was found between HBV infection and DNMT1 up-regulation. These data suggested there is a different mechanism of HBV affected by DNMTs expression in diverse ethnic populations although additional studies with larger sample sizes are required to confirm our findings.
According to the results of both immuno-histo-chemistry and real time RT-PCR, progressive increases in DNMT1 expression may be accompanied by hepatocarcinogenesis from the precancerous stage to the malignant progression of HCC. Immunohistochemical analysis of DNMT1 in biopsy specimens obtained for diagnostic purposes and/or surgically resected materials may show that DNMT1 is a biologic predictor of both HCC recurrence and poor prognosis in HCC patients. To address whether DNMT1 overexpression plays an important role in hepatocellular carcinogenesis in Chinese subjects and to evaluate DNMT1 knockdown strategies for cancer therapy, it is necessary to deplete DNMT1 in the HCC. To avoid the problem of DNMT1 siRNA not being sufficient to inhibit cell growth due to the short-term inhibition of DNMT1 expression, we employed the RNAi technique to knockdown DNMT1 expression in an HCC cell line and assessed tumor cell growth in transiently transfected and stably expressed DNMT1 siRNA cell lines, respectively. Fortunately, we observed that depletion of DNA methyltransferase 1 mediates growth suppression in the HCC cell line SMMC-7721. In order to explore whether this inhibition of DNA replication reflects a distinct alteration in cell cycle kinetics, similar to the DNA damage checkpoints that trigger arrest at distinct phases of the cell cycle[27], we detected PCNA in the treated cells, and found there were no significant changes in PCNA expression in treated cells and controls. However, DNMT1 knockdown inhibited cell growth and induced cell apoptosis in the SMMC-7721 cell line. With a view to the results of cDNA microarray[16], PDCD4, an over-expression apoptosis gene, may contribute to the apoptosis rate in SMMC-7721 cells treated by siRNA. PDCD4 is a proapoptotic molecule involved in TGF-beta1-induced apoptosis in human HCC cells, and a possible tumor suppressor in hepatocarcinogenesis[28–30]. These results provide a rationale for the development of a DNMT1-targeted strategy as an effective epigenetic cancer therapy.
Hepatocellular carcinoma (HCC) is the fourth or fifth largest cause of cancer-related death worldwide. The aberration of DNA methylation (DNMT), which is catalyzed by DNA methyltransferases, is common in HCC. DNMT1 is the main and maintenance methyltransferase in mammals. DNMT1 may play an important role during hepatocarcinogenesis. However, the relationship between abnormal DNMT1 expression and hepatocellular carcinogenesis is still to be elucidated, especially in a Chinese population.
Overexpression of DNMTs is a common event in nearly all malignancies, and DNMT1 may play an important role during hepatocarcinogenesis. In the present study, the authors demonstrated that the overexpression of DNMT1 is correlated significantly with poor tumor differentiation in HCC. Inhibition of DNMT1 through siRNA affected the proliferation ability of an HCC cell line.
Recently, most reports focus on the overexpression of DNMT1 and DNA methylation in tumors. However, abnormal expression of DNMT1 may be involved in tumorigenesis, especially in HBV-related HCC. RNA interference is a direct and efficient way to suppress the target gene in a gene function study. This is the first study to report that inhibition of DNMT1 via RNAi suppressed cell proliferation and induced cell apoptosis in an HCC cell line.
This study implied that DNMT1 maybe considered as a target for HCC therapy. The study also gives a general and better understanding of tumor cell biology and the potential epigenetic mechanism of hepatocellular carcinogenesis.
DNMTs are crucial components of DNA methylation. DNMT1 is the major and best-known DNMT in somatic cells. DNMT1 overexpression correlated significantly with poorer tumor differentiation, but not with the phenotype of the cancer cells, in several tumors. DNMT1 protein might have a more direct and immediate effect on the state of cellular growth and transformation.
The manuscript by Hong et al shows an increased expression of DNMT1 in hepatocellular carcinoma tissue samples and cell lines compared to controls. The increase was correlated with the degree of differentiation. Knockdown of DNMT1 expression in HCC cell lines decreased the number of cells at the end of the experiment. This did not correlate with decreased proliferating cell nuclear antigen (PCNA) expression but was associated with an increase in apoptosis and the expression of apoptosis-related gene PDCD4. For the most part the data is convincing and the experiments are well carried out.
| 2. | McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097-5102. |
| 3. | Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338-342. |
| 4. | Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395-2402. |
| 5. | Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219-220. |
| 6. | Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908-918. |
| 7. | Kimura F, Seifert HH, Florl AR, Santourlidis S, Steinhoff C, Swiatkowski S, Mahotka C, Gerharz CD, Schulz WA. Decrease of DNA methyltransferase 1 expression relative to cell proliferation in transitional cell carcinoma. Int J Cancer. 2003;104:568-578. |
| 8. | Sun L, Hui AM, Kanai Y, Sakamoto M, Hirohashi S. Increased DNA methyltransferase expression is associated with an early stage of human hepatocarcinogenesis. Jpn J Cancer Res. 1997;88:1165-1170. |
| 9. | Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561-568. |
| 10. | Choi MS, Shim YH, Hwa JY, Lee SK, Ro JY, Kim JS, Yu E. Expression of DNA methyltransferases in multistep hepatocarcinogenesis. Hum Pathol. 2003;34:11-17. |
| 11. | Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer. 2001;91:205-212. |
| 12. | Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068-1070. |
| 13. | Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168-174. |
| 14. | Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689-699. |
| 15. | Yasui H, Hino O, Ohtake K, Machinami R, Kitagawa T. Clonal growth of hepatitis B virus-integrated hepatocytes in cirrhotic liver nodules. Cancer Res. 1992;52:6810-6814. |
| 16. | Fan H, Zhao Z, Quan Y, Xu J, Zhang J, Xie W. DNA methyltransferase 1 knockdown induces silenced CDH1 gene reexpression by demethylation of methylated CpG in hepatocellular carcinoma cell line SMMC-7721. Eur J Gastroenterol Hepatol. 2007;19:952-961. |
| 17. | Sato M, Horio Y, Sekido Y, Minna JD, Shimokata K, Hasegawa Y. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene. 2002;21:4822-4829. |
| 18. | Tsuda H, Hirohashi S, Shimosato Y, Terada M, Hasegawa H. Clonal origin of atypical adenomatous hyperplasia of the liver and clonal identity with hepatocellular carcinoma. Gastroenterology. 1988;95:1664-1666. |
| 19. | Liao X, Siu MK, Chan KY, Wong ES, Ngan HY, Chan QK, Li AS, Khoo US, Cheung AN. Hypermethylation of RAS effector related genes and DNA methyltransferase 1 expression in endometrial carcinogenesis. Int J Cancer. 2008;123:296-302. |
| 20. | Park HJ, Yu E, Shim YH. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2006;233:271-278. |
| 21. | Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-687. |
| 22. | Peng DF, Kanai Y, Sawada M, Ushijima S, Hiraoka N, Kitazawa S, Hirohashi S. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27:1160-1168. |
| 23. | Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302-18310. |
| 24. | Nakagawa T, Kanai Y, Saito Y, Kitamura T, Kakizoe T, Hirohashi S. Increased DNA methyltransferase 1 protein expression in human transitional cell carcinoma of the bladder. J Urol. 2003;170:2463-2466. |
| 25. | Li HP, Leu YW, Chang YS. Epigenetic changes in virus-associated human cancers. Cell Res. 2005;15:262-271. |
| 26. | Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476-1494. |
| 27. | Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738-747. |
| 28. | Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101-6112. |