Published online Feb 14, 2009. doi: 10.3748/wjg.15.705
Revised: January 5, 2009
Accepted: January 12, 2009
Published online: February 14, 2009
AIM: To examine the effects of ethanol-induced proteasome inhibition, and the effects of proteasome inhibition in the regulation of epigenetic mechanisms.
METHODS: Rats were fed ethanol for 1 mo using the Tsukamoto-French model and were compared to rats given the proteasome inhibitor PS-341 (Bortezomib, Velcade™) by intraperitoneal injection. Microarray analysis and real time PCR were performed and proteasome activity assays and Western blot analysis were performed using isolated nuclei.
RESULTS: Chronic ethanol feeding caused a significant inhibition of the ubiquitin proteasome pathway in the nucleus, which led to changes in the turnover of transcriptional factors, histone-modifying enzymes, and, therefore, affected epigenetic mechanisms. Chronic ethanol feeding was related to an increase in histone acetylation, and it is hypothesized that the proteasome proteolytic activity regulated histone modifications by controlling the stability of histone modifying enzymes, and, therefore, regulated the chromatin structure, allowing easy access to chromatin by RNA polymerase, and, thus, proper gene expression. Proteasome inhibition by PS-341 increased histone acetylation similar to chronic ethanol feeding. In addition, proteasome inhibition caused dramatic changes in hepatic remethylation reactions as there was a significant decrease in the enzymes responsible for the regeneration of S-adenosylmethionine, and, in particular, a significant decrease in the betaine-homocysteine methyltransferase enzyme. This suggested that hypomethylation was associated with proteasome inhibition, as indicated by the decrease in histone methylation.
CONCLUSION: The role of proteasome inhibition in regulating epigenetic mechanisms, and its link to liver injury in alcoholic liver disease, is thus a promising approach to study liver injury due to chronic ethanol consumption.
- Citation: Oliva J, Dedes J, Li J, French SW, Bardag-Gorce F. Epigenetics of proteasome inhibition in the liver of rats fed ethanol chronically. World J Gastroenterol 2009; 15(6): 705-712
- URL: https://www.wjgnet.com/1007-9327/full/v15/i6/705.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.705
A growing body of evidence indicates that specific histone modifications and modifying enzymes play essential roles in both global and tissue-specific chromatin organization[1]. Histone modifications, such as acetylation and methylation, play important roles in the regulation of gene expression, and impact many fundamental biological processes (i.e. in the cell cycle and cell proliferation). These modifications represent the inheritable epigenetic memory, which is transmitted with high fidelity to future cell generations.
Acetylation of histones is a major factor in the regulation of chromatin remodeling and gene transcription. A sophisticated orchestra of proteins, such as histone acetyltransferases [HATs, gene activation (acetylation)], and histone deacetylases (HDACs), are required to regulate epigenetic mechanisms[2]. Acetylation of histone and non-histone protein is thus emerging as a central process in transcriptional activation, since nuclear HATs act as transcription co-activators, which have been shown to acetylate different transcriptions (i.e. p53, β-catenin, MyoD, SREBP-1)[3].
Alcohol consumption affects epigenetic mechanisms, and causes an increase in histone lysine acetylation, which has been associated with an increase in gene expression that may contribute to uncontrolled transcription[3]. These modifications are the results of changes in the activities of specific modifying enzymes that play essential roles in chromatin structure and specific gene expression. It has previously been shown that chronic ethanol feeding affects epigenetic mechanisms by causing an increase in the stability of histone-modifying enzymes, and thus, in histone modifications[45]. However, little is known about the mechanisms that control the life span of histone-modifying enzymes. For example, the consequence of proteasome inhibition in the nucleus, and its effects on epigenetic mechanisms, have not yet been investigated, and there is very little evidence on proteasome involvement in the turnover of the transcriptional factor and histone-modifying enzymes that regulate epigenetic mechanisms.
Our hypothesis is that proteasome inhibition, induced by ethanol feeding, is associated with histone modification, and is involved in the regulation of histone-modifying enzymes, such as the HAT p300.
The present study demonstrates the role of proteasome activity in epigenetic mechanisms, which significantly contribute to liver injury due to chronic ethanol feeding. Proteasome proteolytic activity regulates histone modifications by regulating the recruitment and stability of histone-modifying enzymes in the nucleus, and, therefore, regulates the chromatin structure, allowing easy access to chromatin by RNA polymerase and enhanced gene expression. The proteasome activity is also believed[6] to be critical for the expression of certain genes, such as those of the enzymes responsible for hepatic transmethylation reactions. In this study, microarray analysis showed up-regulation and down-regulation of a large number of genes, indicating that proteasome activity is essential for up-regulation, as well as down-regulation of specific gene expression. Proteasome inhibition caused a decrease in the gene expression of several enzymes involved in methionine metabolism, particularly betaine-homocysteine methyltransferase (BHMT), which was significantly down-regulated when the proteasome was inhibited[7]. These results indicated that DNA and histone methylation, which play important roles in the regulation of gene silencing, may be affected by proteasome inhibition, and, therefore, may impact many fundamental biological processes.
Previous reports have shown that gene expression changes were numerous at high levels of ethanol, when compared to their pair-fed controls. 1300 genes were changed[8–10]. Similarly, a preliminary microarray analysis of the livers of rats given the proteasome inhibitor PS-341, has shown marked changes in gene expression. Thus, a question arose: which mechanism is involved in this large number of gene expression changes? We believe that this mechanism is epigenetic in nature.
We believe that these modifications in gene expression are the result of a decrease in the activity of the proteasome in the nucleus, which would, for instance, increase the p300 HAT level and activity. Since p300 is responsible for a broad range of gene regulation, the activation of p300 acetylation in histones will increase and activate the expression of a large number of genes in a nonspecific and reversible manner.
The role of the proteasome in regulating histone methylation is also critical because the expression of several genes was changed when the proteasome was inhibited. We have previously demonstrated that proteasome inhibition causes a downregulation in the expression of several genes[8], particularly the gene involved in the remethylation pathway. The study of this remethylation, particularly, the reactional mechanism that regenerates S-adenosylmethionine (SAMe), the major methyl donor, is essential because this is the system that transfers the methyl group to DNA, histones and non-histone proteins via the methyltransferases, such as glycine N-methyltransferase (GNMT).
In this study, BHMT gene expression was markedly decreased by proteasome inhibition. BHMT is an essential enzyme in the remethylation pathway, and is involved in the recovery of SAMe. Betaine, a choline derivative which has been used clinically to treat, with some success, patients with methylenetetrahydrofo-late reductase deficiency[1112], acts as a substrate for BHMT, and serves as an alternative methyl donor for remethylation of homocysteine in the liver and kidney[13]. Therefore, betaine supplementation may cover the down-regulation of gene expression induced by proteasome inhibition, and correct the deregulation of hepatic transmethylation reactions due to the proteasome inhibition-induced decrease in BHMT activity.
Male Wistar rats (Harleco, Hollister, CA, USA), weighing 250-300 g, were fed ethanol using the Tsukamoto-French intragastric model[1415]. PS-341 was administered intraperitoneally, 24 h before sacrifice[1617]. The rats were maintained according to the Guidelines of Animal Care, as described by the National Academy of Sciences and published by the National Institute of Health (1996).
Histones were isolated from the nuclei, according to the method of Umlauf et al[18]. Liver tissues, frozen in isopentane and immersed in liquid nitrogen, were homogenized in a Dounce homogenizer with 10 strokes. Homogenates were centrifuged for 10 min at 6000 ×g. Pellets were resuspended, placed on ice for 10 min, and then centrifuged for 20 min at 9000 ×g on a sucrose cushion. The pellets contained the nuclei. Histones were isolated from the nuclei, according to the method of Shechter et al[19]. Isolated nuclei were mixed with 0.2 mol/L H2SO4, and incubated on a rotator for 30 min at 4°C. Samples were spun in a microcentrifuge at 16000 ×g, for 10 min. Dissolved histones in the supernatant were precipitated with 33% TCA. After acetone wash, histones were dissolved in an appropriate buffer, and further analyses were carried out.
Nuclei were isolated as mentioned above, and 1 &mgr;g of total protein was used. The reaction mixture contained 50 mmol/L Tris–HCl pH 8, 1 mmol/L DTT, and 40 &mgr;mol/L Suc-LLVY-AMC substrate for chymotrypsin-like activity. The mixture was incubated for 30 min at 37°C, and the reaction was then stopped by adding 100 &mgr;mol/L monochloroacetate and 30 mmol/L sodium acetate (pH 4.3). Fluorescence was determined by measuring the release of AMC (λ excitation: 370 nm, λ emission: 430 nm) using a Perkin Elmer LS 30 spectrofluorometer.
Proteins (50 &mgr;g) from isolated nuclei or isolated histones were separated by SDS-PAGE, and transferred to a PVDF membrane (Bio-Rad, Hercules, CA, USA) for 1 h in 25 mmol/L Tris-HCl (pH 8.3), 192 mmol/L glycine and 20% methanol. The membranes were stained using primary antibodies to the antigens. The appropriate species anti-polyclonal and anti-monoclonal HRP-conjugated antibodies were used as secondary antibodies. The membranes were subjected to chemiluminescence detection using luminal, according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Fast frozen rat liver tissue was subjected to microarray analysis. Total liver RNAs were extracted with UltraspecTM RNA Isolation Systemic (Biotecx Laboratories, Houston, TX, USA), and cleaned with Rneasy columns (Qiagen, Valencia, CA, USA). Five micrograms of total RNA were used for preparing biotin-labeled cRNA. Labeled and fragmented cRNA was subsequently hybridized to Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA, USA). Labeling, hybridization, image scanning, and initial data analysis were performed at the Microarray Core at Los Angeles Biomedical Research Institute. Sample preparation and loading, hybridization, staining, and microarray data analysis were then performed[8].
Total liver RNAs were extracted with Trizol Plus RNA Purification Kit (Invitrogen, Carlsbad, CA, USA). Synthesis of cDNAs was performed with 5 &mgr;g total RNA, and 50 ng random hexamer primers using SuperScriptIII RNase H-Reverse Transcriptase (Invitrogen). PCR primers were designed using the Primer Express software (Applied Biosystems, Foster City, CA, USA). The primers for rat p300 are shown in Table 1.
| p300 | XM_576312 | Forward | 5GAGGTCACTGTTCGGGTTGTTC |
| p300 | XM_576312 | Reverse | 5TGGTTCGATATGGAAAAGATTCTG |
| BHMT | NM_030850 | Forward | 5GGGCAGAAGGTCAATGAAGCT |
| BHMT | NM_030850 | Reverse | 5ACCAATGCATCCCCTTCGT |
Quantitative PCR was achieved using the SYBR Green JumpStartTM (Applied Biosystems). Thermal cycling consisted of an initial step at 50°C for 2 min, followed by a denaturation step at 95°C for 10 min, and then 40 cycles at 95°C for 15 s and 60°C for 1 min. A single PCR product was confirmed with the heat dissociation protocol at the end of the PCR cycles. Each data point was repeated three times.
Sense and anti-sense: quantitative PCR was achieved using the SYBR Green JumpStart™ Tag ReadyMix (Sigma, St. Louis, MO, USA) on an ABI PRISM 7700 Sequence Detector System (Applied Biosystems). The thermal cycling consisted of an initial step at 50°C for 2 min, followed by a denaturation step at 95°C for 10 min, then 40 cycles at 95°C for 15 s and 60°C for 1 min. A single PCR product was confirmed with the heat dissociation protocol at the end of the PCR cycles. Quantitative values were obtained from the threshold PCR cycle number (Ct) at which point the increase in signal associated with an exponential growth for the PCR product was detected. The target mRNA abundance in each sample was normalized to its 18S level as ΔCt = Cttarget gene-Ct18S. For each target gene, the highest ΔCt was assigned as ΔCtmax.
Data were obtained from at least three animals in each group. Bars represent mean ± SE. P values were determined by one-way ANOVA and Student-Newman Keuls for multiple group comparisons (Sigma-Stat software, San Francisco, CA, USA). P≤ 0.05 was used to establish significant differences. Correlation of data was done by linear regression analysis using Pearson’s period momentum method.
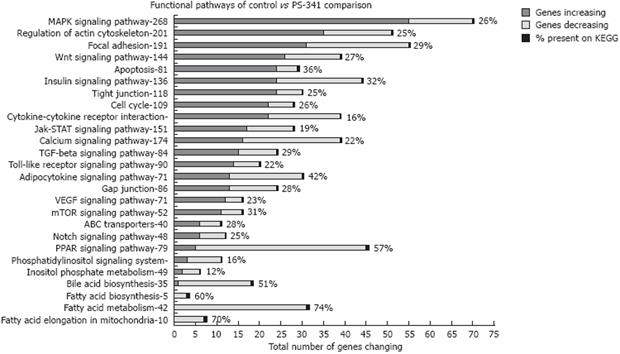
Microarray analysis of liver samples from rats fed ethanol showed that a large number of genes (about 1300) were up-regulated and down-regulated due to chronic ethanol feeding[8]. Microarray analyses of liver samples from rats given PS-341 (Bortezomib, Velcade®) also showed dramatic changes in gene expression (about 2082 genes changed) affecting several functional pathways (Figure 1).
The present study was based on the observation that the inhibition of proteasome, caused by chronic ethanol feeding, participated in the development of liver injury due to ethanol by altering the mechanisms through which normal epigenetic regulation occurs. The consequence of this was a marked change in the gene expression of several functional pathways in liver cells (Figure 1).
Data mining and gene specific pathway clustering showed that, similar to ethanol feeding, several transcriptional factors, such as cell cycle, histone modifying enzymes, and the remethylation pathway, were significantly changed by proteasome inhibition. Proteasome inhibition by PS-341 thus proved to be a powerful tool to investigate the role of proteasome activity in epigenetic mechanisms.
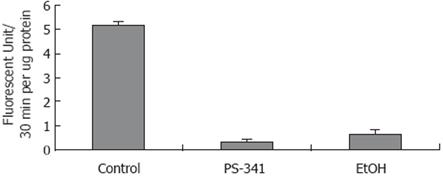
To verify the hypothesis that gene expression changes are regulated by nuclear proteasome activity, where inhibition is caused by chronic ethanol feeding[20], proteasome activity was measured in isolated nuclei from the liver of rats fed ethanol chronically, and from the liver of rats given PS-341. Figure 2 shows that chronic ethanol feeding caused a significant decrease in proteasome chymotrypsin-like activity in isolated liver nuclei.
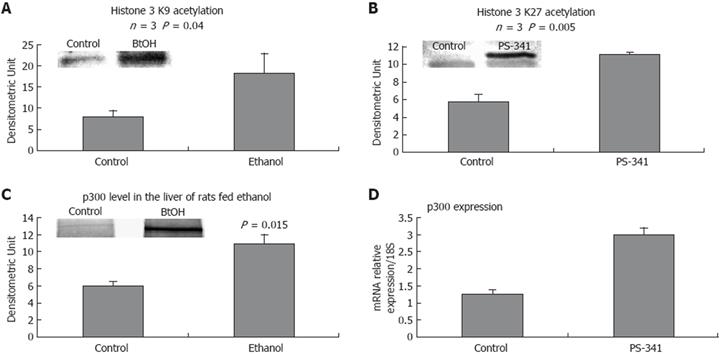
To further investigate the role of proteasome activity in regulating epigenetic mechanisms, histone acetylation was analyzed in the liver of rats given PS-341, and compared to histone acetylation in the liver of rats fed ethanol chronically. Figure 3A shows that acetylated histone 3 lysine 9 (AcH3K9) was increased in the liver of rats fed ethanol, and that acetylated histone 3 lysine 27 (AcH3K27) was increased in the liver of rats given PS-341 (Figure 3B).
Increased acetylation was concomitant with an increase in the level of HAT p300, which was linked to significant proteasome inhibition shown in the liver of rats fed ethanol chronically (Figure 3D), and in the liver of rats given PS-341 (Figure 3C). P300 was increased in the ethanol-isolated nuclear extract, which confirmed a previous report[4]. It is now well established that ethanol feeding increases histone acetylation[45], which correlates with an increase in the acetyltransferase CBP/p300, and a decrease in Sirt1 activity[321]. Under our experimental conditions, Sirt1 gene expression and protein level were up-regulated[22], or showed no significant changes[4]. However, the activity of the enzyme may be decreased due to the low level of NAD+, a Sirt1 cofactor[23].
The increased level of histone acetylation substantiated the increase in p300 activity in the liver of rats fed ethanol. In addition, proteasome inhibition by PS-341 caused a significant increase in histone acetylation, which substantiated our hypothesis that ethanol induced-histone acetylation is associated with proteasome inhibition. The level of p300 was increased in the nucleus when ethanol was fed chronically, and may also have accumulated due to proteasome proteolysis slowdown in the nucleus.
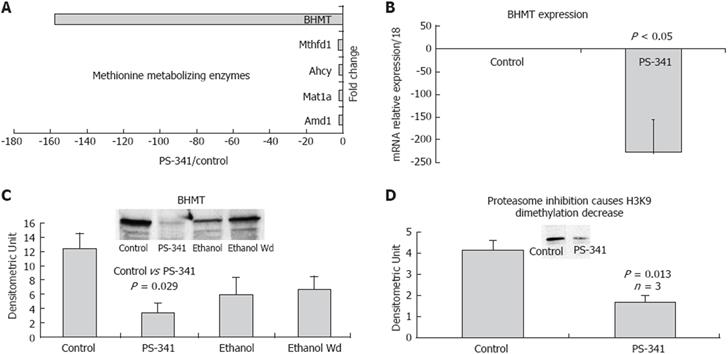
Proteasome activity has also been found to be involved in the regulation of remethylation enzyme gene expression, thus affecting DNA and histone methylation when it is inhibited. Microarray data mining showed a down-regulation in gene expression of the remethylation enzymes when the proteasome was inhibited (Figure 4A), especially BHMT, which reconstitutes methionine, the major element in the methylation pathway (Figure 4B). Mat1a, Adm (S-adenosylmethionine decarboxylase), and Ahcy (S-adenosylhomocysteine hydrolase) were also down-regulated by proteasome inhibition (Figure 4A), indicating the role of proteasome activity in the methionine-metabolizing enzymes system. Figure 4C shows that proteasome inhibition significantly decreased the protein level of BHMT in the liver of rats given PS-341, and, to a lower extent, in rats subjected to chronic ethanol feeding. These findings demonstrated the role of the proteasome in regulating gene expression in the remethylation pathway, and the role of proteasome activity in the cellular remethylation pathway. Western blot analysis showed a significant decrease in H3K9 dimethylation in the liver of rats given PS-341. A decrease in H3K9 dimethylation (Figure 4D) was also observed in ethanol-treated cells[24], suggesting similar effects of proteasome inhibition and ethanol feeding. These results showed for the first time the role of proteasome activity in regulating the mechanisms of cellular remethylation, and is a promising approach in chemotherapy regimens which use proteasome inhibitor as an anti tumor drug.
Several studies have shown that the inhibition of the ubiquitin-proteasome pathway is a pathobiological mechanism associated with the development of liver disease, especially alcoholic liver disease[2526]. We believe that ethanol-induced inhibition of proteasome activity may play a significant role in the deregulation of epigenetic mechanisms and the mechanism related to liver injury in alcoholic liver disease (ALD).
Many cellular signaling pathways are controlled by selective proteolysis of key regulatory proteins via the ubiquitin-proteasome system[41227]. Proteasome is involved in RNA polymerase II degradation[28], which is a key step in controlling the transcriptional mechanism, preventing uncontrolled transcription that may occur when ethanol metabolism generates oxidative stress, which causes DNA damage[29]. Reports have shown that proteasome dysfunction leads to apoptotic death of hepatocytes and sensitization to tumor necrosis factor (TNF) cytotoxicity, leading to direct hepatic injury[25]. Although it is now evident that inhibition of cytoplasmic proteasome function is consistently shown in ALD models, the way in which proteasome dysfunction may enhance hepatotoxicity is not well defined. In addition, the effects of ethanol feeding on the activity of nuclear proteasome and the consequences of proteasome inhibition on changes in epigenetic mechanisms have not yet been demonstrated. Most importantly, our previous investigations have repeatedly shown that chronic ethanol feeding causes significant proteasome inhibition in the cell[3031]. The ubiquitin-proteasome pathway is the cellular proteolytic pathway dedicated to controlling protein stability, and understanding the link between the ubiquitin-proteasome pathway and the histone-modifying machinery will define the link between proteasome proteolytic activity, epigenetic mechanisms, and the effect of toxic substances, such as ethanol and its metabolism generated end products, in the regulation and control of epigenetic mechanisms.
As predicted, our results showed an increase in acetylation of H3K9 in the liver nuclear extracts of rats fed ethanol chronically. This increase was associated with an increase in HAT p300, which confirmed a previous report[4]. In addition, p300 activation and histone acetylation were also obtained when proteasome activity was inhibited using PS-341, which supported the role of proteasome activity in regulating the stability of p300 in the nucleus and therefore supported the role of proteasome in regulating the acetylation mechanisms and thus gene expression. These results corroborate the findings of Marcu et al[32], which showed that p300 is a proteasome substrate, and that p300 is accumulated when the proteasome is inhibited.
The modification of histones mirrors the sophisticated protein machinery that controls gene expression and regulates transcription. Therefore, it would be naïve to suggest that, for instance, only H3K9 acetylation explained all the changes in the observed gene expression[11]. The balance between all histone lysine residue modifications accounted for the pattern of gene expression and further histone modifications are certainly involved in defining specific gene expression.
Histone methylation also plays a critical role in regulating gene expression and transcription. The cellular remethylation pathway is the major player in the methylation mechanism, because it produces the methyl donor SAMe. It is well known that chronic ethanol ingestion causes a serious deregulation of this pathway. Methionine adenosyltransferase 1-alpha, which is responsible for the conversion of methionine to SAMe, is decreased in the liver of rats fed ethanol[33]. Since SAMe is thus decreased, the level of DNA and protein methylation is decreased[34], and the level of homocysteine is increased[35]. The ratio SAMe/SAH (S-adenosylhomocysteine) is critical in the cell because it controls most of the methyltransferase activity. BHMT, which hydrolyzes betaine, helps remove SAH and homocysteine with subsequent regeneration of the methyl group and a reduction in the level of SAH. Our results show that BHMT was significantly down-regulated when the proteasome activity was inhibited, either by chronic ethanol feeding or by treatment with a proteasome inhibitor. In addition, there was a decrease in histone methylation, reflecting the role of proteasome activity in regulating the methylation mechanisms in the liver cell. Proteasome inhibition plays a critical role in regulating the gene expression of key enzymes in the remethylation pathway, such as BHMT.
Alcohol ingestion causes alterations in several cellular mechanisms, and leads to inflammation, apoptosis, and fibrosis. These phenomena are associated with significant changes in epigenetic mechanisms and with a subsequent liver cell memory. The ubiquitin-proteasome pathway is a vital cellular pathway which undergoes dysfunction due to chronic ethanol consumption.
Although inhibition of proteasome function has been widely reported in models of alcoholic liver disease (ALD), why proteasome dysfunction may enhance hepatotoxicity is not well defined. In addition, there is no evidence of the effect of ethanol feeding on the activity of nuclear proteasome and the consequences of proteasome inhibition in epigenetic mechanisms and DNA repair.
The present study focused on the role of proteasome activity in gene expression and the effects of proteasome inhibition in changing epigenetic mechanisms. The model used to study the consequences of proteasome inhibition due to chronic intragastric tube ethanol administration was proteasome inhibition by PS-341, a dipeptide boronic acid currently used in clinical trials as an anti tumor drug, and is associated with profound side effects. Inhibition of proteasome activity occurred in the nucleus of liver cells taken from rats fed ethanol chronically and from rats given PS-341. This inhibition caused changes in the turnover of transcriptional factors, histone modifying enzymes, and, therefore, affected epigenetic mechanisms. Histone acetylation was increased following both treatments and gene expression was changed. Identification of DNA and histone modifications was critical in regulating gene expression, especially genes involved in the cell cycle and apoptosis, and those involved in the metabolism of ethanol. In addition, proteasome inhibition has been shown to significantly affect the hepatic remethylation pathway. In particular, proteasome inhibition caused a decrease in gene expression of the enzyme betaine-homocysteine methyltransferase (BHMT), which is involved in the recovery of s-adenosylmethionine (SAMe). Histone methylation was also decreased when the proteasome was inhibited suggesting that hypomethylation was associated with the decrease in proteasome activity.
The phenomenon of hypomethylation is currently corrected by diet supplementation with a methyl donor (betaine) to redirect methionine/homocysteine metabolism toward recovery of methionine, and SAMe regeneration.
Chronic ethanol feeding causes proteasome inhibition, which leads to cellular dysfunction including the accumulation of damaged proteins which form Mallory bodies in the liver of severe alcoholic patients, immune response deficiency, cell cycle deregulation and incorrect gene expression. The mechanism of incorrect gene expression was the focus of this study, particularly the consequences of ethanol induced-proteasome inhibition in the nucleus and the role played by proteasome in regulating epigenetic mechanisms.
The impact of this study is great because when epigenetic mechanisms associated with proteasome inhibition are fully identified, a therapeutic approach will be initiated to counteract the ethanol induced-epigenetic changes, and prevent any cellular memory for future cell generations.
| 1. | Lin W, Dent SY. Functions of histone-modifying enzymes in development. Curr Opin Genet Dev. 2006;16:137-142. |
| 2. | Bronner C, Chataigneau T, Schini-Kerth VB, Landry Y. The "Epigenetic Code Replication Machinery", ECREM: a promising drugable target of the epigenetic cell memory. Curr Med Chem. 2007;14:2629-2641. |
| 3. | You M, Liang X, Ajmo JM, Ness GC. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892-G898. |
| 4. | Bardag-Gorce F, French BA, Joyce M, Baires M, Montgomery RO, Li J, French S. Histone acetyltransferase p300 modulates gene expression in an epigenetic manner at high blood alcohol levels. Exp Mol Pathol. 2007;82:197-202. |
| 5. | Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126-132. |
| 6. | Kinyamu HK, Archer TK. Proteasome activity modulates chromatin modifications and RNA polymerase II phosphorylation to enhance glucocorticoid receptor-mediated transcription. Mol Cell Biol. 2007;27:4891-4904. |
| 7. | Dedes J, Li J, Bardag-Gorce F. Chromatin remodeling is regulated by the ubiquitin proteasome pathway. FASEB J. 2008;22:lb194. |
| 8. | Bardag-Gorce F, French BA, Dedes J, Li J, French SW. Gene expression patterns of the liver in response to alcohol: in vivo and in vitro models compared. Exp Mol Pathol. 2006;80:241-251. |
| 9. | French BA, Dedes J, Bardag-Gorce F, Li J, Wilson L, Fu P, Nan L, French SW. Microarray analysis of gene expression in the liver during the urinary ethanol cycle in rats fed ethanol intragastrically at a constant rate. Exp Mol Pathol. 2005;79:87-94. |
| 10. | Tsukamoto H, French SW, Reidelberger RD, Largman C. Cyclical pattern of blood alcohol levels during continuous intragastric ethanol infusion in rats. Alcohol Clin Exp Res. 1985;9:31-37. |
| 11. | Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163-176. |
| 12. | Bardag-Gorce F, Oliva J, Villegas J, Fraley S, Amidi F, Li J, Dedes J, French B, French SW. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;84:113-121. |
| 13. | Fuchs SY. The role of ubiquitin-proteasome pathway in oncogenic signaling. Cancer Biol Ther. 2002;1:337-341. |
| 14. | Li J, Nguyen V, French BA, Parlow AF, Su GL, Fu P, Yuan QX, French SW. Mechanism of the alcohol cyclic pattern: role of the hypothalamic-pituitary-thyroid axis. Am J Physiol Gastrointest Liver Physiol. 2000;279:G118-G125. |
| 15. | French SW. Intragastric ethanol infusion model for cellular and molecular studies of alcoholic liver disease. J Biomed Sci. 2001;8:20-27. |
| 16. | Bardag-Gorce F, Li J, French BA, French SW. Ethanol withdrawal induced CYP2E1 degradation in vivo, blocked by proteasomal inhibitor PS-341. Free Radic Biol Med. 2002;32:17-21. |
| 17. | Bardag-Gorce F, Francis T, Nan L, Li J, He Lue Y, French BA, French SW. Modifications in P62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci. 2005;77:2594-2602. |
| 18. | Umlauf D, Goto Y, Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol. 2004;287:99-120. |
| 19. | Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445-1457. |
| 20. | Bardag-Gorce F, French BA, Nan L, Song H, Nguyen SK, Yong H, Dede J, French SW. CYP2E1 induced by ethanol causes oxidative stress, proteasome inhibition and cytokeratin aggresome (Mallory body-like) formation. Exp Mol Pathol. 2006;81:191-201. |
| 21. | You M, Cao Q, Liang X, Ajmo JM, Ness GC. Mammalian sirtuin 1 is involved in the protective action of dietary saturated fat against alcoholic fatty liver in mice. J Nutr. 2008;138:497-501. |
| 22. | Oliva J, French BA, Li J, Bardag-Gorce F, Fu P, French SW. Sirt1 is involved in energy metabolism: the role of chronic ethanol feeding and resveratrol. Exp Mol Pathol. 2008;85:155-159. |
| 23. | Bardag-Gorce F, French BA, Li J, Riley NE, Yuan QX, Valinluck V, Fu P, Ingelman-Sundberg M, Yoon S, French SW. The importance of cycling of blood alcohol levels in the pathogenesis of experimental alcoholic liver disease in rats. Gastroenterology. 2002;123:325-335. |
| 24. | Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81:979-987. |
| 25. | Donohue TM Jr. The ubiquitin-proteasome system and its role in ethanol-induced disorders. Addict Biol. 2002;7:15-28. |
| 26. | French SW, Mayer RJ, Bardag-Gorce F, Ingelman-Sundberg M, Rouach H, Neve And E, Higashitsuji H. The ubiquitin-proteasome 26s pathway in liver cell protein turnover: effect of ethanol and drugs. Alcohol Clin Exp Res. 2001;25:225S-229S. |
| 27. | Starkova NN, Koroleva EP, Rotanova TV. [Intracellular proteolysis: signals of selective protein degradation]. Bioorg Khim. 2000;26:83-96. |
| 28. | Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375-1388. |
| 29. | Ribar B, Prakash L, Prakash S. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3999-4005. |
| 30. | Bardag-Gorce F, Yuan QX, Li J, French BA, Fang C, Ingelman-Sundberg M, French SW. The effect of ethanol-induced cytochrome p4502E1 on the inhibition of proteasome activity by alcohol. Biochem Biophys Res Commun. 2000;279:23-29. |
| 31. | Bardag-Gorce F, Venkatesh R, Li J, French BA, French SW. Hyperphosphorylation of rat liver proteasome subunits: the effects of ethanol and okadaic acid are compared. Life Sci. 2004;75:585-597. |
| 32. | Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, Neckers L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169-174. |
| 33. | Song Z, Zhou Z, Song M, Uriarte S, Chen T, Deaciuc I, McClain CJ. Alcohol-induced S-adenosylhomocysteine accumulation in the liver sensitizes to TNF hepatotoxicity: possible involvement of mitochondrial S-adenosylmethionine transport. Biochem Pharmacol. 2007;74:521-531. |
| 34. | Kharbanda KK, Mailliard ME, Baldwin CR, Beckenhauer HC, Sorrell MF, Tuma DJ. Betaine attenuates alcoholic steatosis by restoring phosphatidylcholine generation via the phosphatidylethanolamine methyltransferase pathway. J Hepatol. 2007;46:314-321. |
| 35. | Purohit V, Abdelmalek MF, Barve S, Benevenga NJ, Halsted CH, Kaplowitz N, Kharbanda KK, Liu QY, Lu SC, McClain CJ. Role of S-adenosylmethionine, folate, and betaine in the treatment of alcoholic liver disease: summary of a symposium. Am J Clin Nutr. 2007;86:14-24. |