Published online Feb 21, 2008. doi: 10.3748/wjg.14.1067
Revised: November 22, 2007
Published online: February 21, 2008
AIM: To investigate the ability of Lactic acid bacteria (LAB) to modulate inflammatory reaction in human intestinal cell lines (Caco-2, HT-29 and HCT116). Different strains of LAB isolated from new born infants and fermented milk, together with the strains obtained from culture collections were tested.
METHODS: LABs were treated with human intestinal cell lines. ELISA was used to detect IL-8 and TGF-β protein secretion. Cytokines and Toll like receptors (TLRs) gene expression were assessed using RT-PCR. Conditional medium, sonicated bacteria and UV killed bacteria were used to find the effecter molecules on the bacteria. Carbohydrate oxidation and protein digestion were applied to figure out the molecules’ residues. Adhesion assays were further carried out.
RESULTS: It was found that Enterococcus faecalis is the main immune modulator among the LABs by downregulation of IL-8 secretion and upregulation of TGF-β. Strikingly, the effect was only observed in four strains of E. faecalis out of the 27 isolated and tested. This implies strain dependent immunomodulation in the host. In addition, E. faecalis may regulate inflammatory responses through TLR3, TLR4, TLR9 and TRAF6. Carbohydrates on the bacterial cell surface are involved in both its adhesion to intestinal cells and regulation of inflammatory responses in the host.
CONCLUSION: These data provide a case for the modulation of intestinal mucosal immunity in which specific strains of E. faecalis have uniquely evolved to maintain colonic homeostasis and regulate inflammatory responses.
-
Citation: Wang S, Ng LHM, Chow WL, Lee YK. Infant intestinal
Enterococcus faecalis down-regulates inflammatory responses in human intestinal cell lines. World J Gastroenterol 2008; 14(7): 1067-1076 - URL: https://www.wjgnet.com/1007-9327/full/v14/i7/1067.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.1067
The role of gastrointestinal microflora, such as several strains of lactic acid bacteria (LAB) in priming the immune system during ontogeny to limit allergy and chronic inflammatory responses has been brought to attention in recent years[12]. Clinical and experimental studies have put forward that gastrointestinal microflora is essential for maintenance of intestinal homeostasis, and their influence may extend beyond the gut, modifying systemic immunity. Studies using intestinal epithelial cell (IEC) lines have suggested that commensal bacteria may “program” IECs to prevent or down-modulate pro-inflammatory responses by interfering with Toll like receptors (TLRs) expression[34]. At least 13 TLRs have been identified in humans. Among all the 13 members of TLRs, TLR3, TLR5, TLR9 are mainly expressed by epithelial cells. They could be activated by pathogens leading to the initiation of inflammation by secreting pro-inflammatory cytokines such as Interleukin-8 (IL-8) and Tumor necrosis factor (TNF)-α[5].
The population of microbes harbored in the human intestine is dynamic and varies with age. It was reported that infants who developed allergy were often found to possess less enterococci in the intestines than healthy infants during the first month of life[6–8]. Enterococcus faecalis is a major commensal gastrointestinal flora in humans[910]. So far, it is not clear if the depletion of enterococci in allergic children is a cause or effect of allergy. Although there are some reports that E. faecalis caused infections in immune deficient mice[11], many studies also showed that E. faecalis could suppress the proliferation of intestinal pathogens and thus may prevent infection and the induction of inflammation[12–15]. Another study showed that E. faecalis lysate could relieve the clinical symptoms of Japanese cedar pollinosis[16]. However, the role and effect of E. faecalis in inflammation reactions are not clear. It is the intention of this study to shed light on the role and mechanisms of anti-inflammatory effect of enterococci. This knowledge could provide a therapeutic window and development of treatments or prophylaxis of intestinal infectious, inflammatory and allergic reactions and perhaps carcinogenesis.
In the present study, the anti-inflammatory effect of human intestinal LABs isolated from infants and probiotic bacteria with alleged anti-allergic properties were investigated in human intestinal cell lines (Caco-2, HT-29 and HCT116). It is demonstrated that E. faecalis is the main immune modulator among the intestinal LABs. However, the effect is limited to only a few strains of E. faecalis. Carbohydrate components on bacterial cell walls were the regulators of the inflammatory responses in the host.
Caco-2, HT-29 and HCT116 cells were obtained from American Type Culture Collection (Manassas, VA). Caco-2 and HT-29 are human colonic adenocarcinoma cells. Differentiated Caco-2 are enterocyte-like cells[17] whereas HCT116 are human colon carcinoma cells. All cells were cultured at 37°C in a 5% CO2 atmosphere. Caco-2 cells were cultured in MEM (GIBCO, Grand Island, NY) containing 20% fetal bovine serum (Bioclot, Germany) and 1% nonessential amino acids (GIBCO). HT-29 and HCT116 cells were cultured in DMEM (GIBCO) containing 10% fetal bovine serum (Bioclot, Germany). All culture media were supplemented with 100 U/mL penicillin and 100 &mgr;g/mL streptomycin (GIBCO).
The origin of the bacterial strains used in the experiment is indicated in Table 1. Lactobacilli were stored in De Man, ROGOSA and SHARPE (MRS) agar (Merck, Darmstadt, Germany) plus 25% glycerol, at -80°C and grown in MRS broth overnight at 37°C without shaking. Enterococci were stored in Tryptone Soy broth (TSB) (Oxoid, England) plus 25% glycerol at -80°C and grown in TSB overnight at 37°C without shaking. Streptococcus thermophilus NCIMB 10387 (10387) was grown on M17 medium (Oxoid, England). Salmonella typhimurium 14028 (Salm) was maintained on Luria-Bertani (LB) Agar (BBL, Cockeyville, MD, USA). The freshly cultured bacterial strains were further sub-cultured in a 250 mL conical flask with shaking at 120 r/min overnight. The bacteria were then pelleted by centrifugation, washed using serum-free medium three times and then resuspended in complete medium for use.
| Lactic acid bacteria (LAB) | Origin | Remark |
| Bifidobacterium adolescentis ATCC15703 | Infant intestine | - |
| B. brevi ATCC15700 | Infant intestine | - |
| B. dentium scardori & crociani ATCC27534 | Dental carries | - |
| B. longum ATCC15707 | Adult intestine | - |
| Enterococcus faecalis EC1 | Infant feces | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| Age: 1 mo | ||
| Enterococcus faecalis EC3 | Infant feces | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| Age: 3 d | ||
| Enterococcus faecalis EC15 | Infant feces | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| Age: 3 d | ||
| Enterococcus faecalis EC16 | Infant feces | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| Age: 3 d | ||
| Other 23 strains of enterococci and lactobacilli | Infant feces | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| Age: 3 d to 1 mo | ||
| Lactobacillus acidophilus ATCC4356 | Human | - |
| L. brevis K7 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. brevis T1 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. brevis T6 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. casei ATCC11578 | Oral | - |
| L. casei IS7257 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. casei subsp. casei NCIMB11970 | Cheese | - |
| L. casei Shirota | Human | Commercial probiotic |
| L. delbrueckii subsp. bulgaricus NCIMB11778 | Bulgarian yogurt | Commercial probiotic |
| L. delbrueckii bulgaricus D1 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. lactis subsp. lactis B4 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. lactis subsp. lactis B9 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. lactis subsp. lactis B12 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. lactis subsp. lactis K5 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. lactis subsp. lactis K6 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. lactis subsp. lactis K7 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. paracasei LP33 | Fermented milk | Commercial probiotic |
| Lactobacillus paracasei Chamyto | Yogurt | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| L. paracasei subsp. paracasei ATCC11974 | Dental carries | - |
| L. paracasei subsp. paracasei NCIMB8001 | - | - |
| L. rhamnusus NCIMB6375 | Oral | |
| L. rhamnusus GG | Human feces | Commercial probiotic |
| L. rhamnosus NCIMB 8690 | - | - |
| Leuconostoc mesentroides K13 | Fermented milk | Identified by phenotypic characterization and sequence analysis of 16S rDNA |
| Streptococcus thermophilus NCIMB10387 | Yogurt | Commercial probiotic |
To obtain a polarized monolayer of intestinal epithelial cells (IECs), 1 × 105 Caco-2 cells were grown in a sterile 24-well plate (Nalge Nunc International, USA) to confluence and then for 2 additional weeks to reach full differentiation and polarization. The culture medium was changed every two days. For HT-29 and HCT116 cell lines, 1 × 105 cells were cultured in sterile 24-well flat-bottom plates (Nalge Nunc International, USA) for 24 h before the infection. For the infection step, Caco-2, HT-29 and HCT116 cells were incubated in fresh medium with bacteria at a multiplicity of infection (MOI) of 100 or 1000 for 6 h. Biological triplicates were done.
After the infection step, Caco-2, HT-29 and HCT116 cells were washed with cold PBS twice. RNAs were then extracted with a RNA extraction kit (Roche Applied Science, Germany) according to the manufacturer’s instruction. Samples were stored at -80°C before assay. The integrity and quality of RNA were determined by running formaldehyde gel. cDNA were synthesized from a template of 1 &mgr;g total RNA using the Superscript III (Invitrogen Life Technologies, UK) according to the manufacturer’s instruction.
PCR were performed using 2 &mgr;L of the cDNA as a template in a final volume of 50 &mgr;L. The standard program used was as follows: denaturation for 2 min at 94°C, 35 cycles of 30 s at 94°C, 1 min at 55°C or 60°C, and 1 min at 72°C, followed by a final elongation for 7 min at 72°C. 0.2 unit of Taq (Invitrogen Life Technologies, UK) was used for one PCR. H2O was used as a negative control and plasmids containing the specific genes were used as a positive control in the PCR step. After electrophoresis in a 1.2% agarose gel containing Ethidium Bromide, PCR products were quantified using the Gene Tool software (Syngene, UK). The primers used and annealing temperatures are described in Table 2.
| Gene name | Forward | Reverse | Annealing temp (°C) | Length (bp) |
| β-actin | GGCGACGAGGCCCAGAGCAAGAGAGGCAT | CGATTTCCCGCTCGGCCGTGGTGGTGAAGC | 55 | 460 |
| IL-4 | AACACAACTGAGAAGGAAACCTTC | GCTCGAACACTTTGAATATTTCTC | 55 | 276 |
| IL-8 | ATGACTTCCAAGCTGGCCGTGGCT | TCTCAGCCCTCTTCAAAAACTTCTC | 55 | 289 |
| IL-10 | ATGCCCCAAGCTGAGAACCAAGACCCA | TCTCAAGGGGCTGGGTCAGCTATCCCA | 55 | 352 |
| TGF-β | TGACAGCAGGGATAACACACT | GTAGGGGCAGGGCCCGAGGCA | 55 | 288 |
| TLR1 | CTATACACCAAGTTGTCAGC | GTCTCCAACTCAGTAAGGTG | 55 | 219 |
| TLR2 | GCCAAAGTCTTGATTGATTGG | TTGAAGTTCTCCAGCTCCTG | 55 | 346 |
| TLR3 | GATCTGTCTCATAATGGCTTG | GACAGATTCCGAATGCTTGTG | 55 | 304 |
| TLR4 | CTTTATCCAACCAGGTGC | GGAATGCTGGAAATCCAG | 50 | 650 |
| TLR5 | TAGCTCCTAATCTGATG | CCATGTGAAGTCTTTGCTGC | 50 | 437 |
| TLR9 | GTCCCCACTTCTCCATG | GGCACAGTCATGATGTTGTTG | 55 | 259 |
| TNF-α | CAGAGGGAAGAGTTCCCCAG | CCTTGGTCTGGTAGGAGACG | 55 | 324 |
| Tollip | CGCGGTACCGCCACCATGGCGACCACCGTCAGC | GCCGGGCCCTGGCTCCTCCCCCATCTG | 60 | 850 |
| TRAF6 | GTCGGTACCGCCACCATGAGTCTGCTAAACTGTGAA | CGCGGGCCCCTATACCCCTGCATCAGTAC | 60 | 1600 |
In this study, the expression of TLR1, TLR2, TLR3, TLR4, TLR5, TLR9, Tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6) and Toll inhibitory protein (Tollip) were measured using RT-PCR. Cytokine gene expression of IL-8, IL-4, IL-10, TGF-β and TNF-α were also detected using this method.
For the tests in this part, we chose only four strains of LAB [E. faecalis (EC1, EC3, EC15, EC16)] to infect HCT116 cells with a MOI of 1000. The cells were co-cultured with bacteria at 37°C with 5% CO2 for 6 h. The supernatants were then harvested for cytokines assay. Unless indicated otherwise all enzymes and chemicals were from Sigma, USA.
Cell wall component and crude cell extract preparation: Before sonication, a protease inhibitor (2 mmol/L phenylmethylsulfonylfluoride (PMSF), 1 mmol/L dithiothreitol (DTT) and 1 mmol/L sodium metavanadate) was added to bacterial suspensions. The bacterial cells were then disrupted by sonication (MSE Soniprep 150, SANYO, Japan) on ice for 10 cycles with 30 s pulses and 1 min rest[18]. Cell debris was pelleted down by centrifugation at 15 000 ×g for 60 min at 0°C. The bacterial culture supernatant was aseptically kept on ice for use. The remaining pellet was resuspended in an appropriate volume of complete DMEM. One mL of each supernatant and 1 mL of each cell debris suspension were added to the cell culture. Biologically triplicates were done on this experiment.
Conditional medium and cell inserts treatment: In order to test for the location of the active components, the cell culture supernatants obtained from “infection of intestinal cells” were harvested sterilely and were then added to the cell culture prepared as in section “infection of intestinal cells”. Supernatants were harvested for further cytokine assay testing. In addition, bacteria themselves were also cultured with the control supernatants obtained from the section “infection of intestinal cells”. All the treated cells were then incubated at 37°C with 5% CO2 for 6 h. The supernatants were then harvested for cytokines assay.
For the cell insert test, HCT116 cells were plated in wells of Transwell® (USA) multiple well plate and incubated at 37°C with 5% CO2 for 24 h. Accompanying inserts were added 1 h before co-culturing. 100 &mgr;L of bacterial suspensions with correct concentration were added to each insert.
Triplicates of each condition were done and supernatants were harvested for the cytokine assay test.
Carbohydrate oxidation and protein digestion of bacterial cell wall test: 4 mL of bacterial cells were pelleted by centrifugation at 15 000 g at 0°C for 10 min. The bacterial cells were washed once in PBS and then suspended in 1.5 mL of one of the following solutions: Solution 1 was 0.05 mol/L sodium m-periodate in buffer A (5 mol/L LiCl in water; 0.1 mol/L citrate-phosphate-0.1 mol/L NaCl) with a final pH 4.5. Solution 2 was 0.05 mol/L sodium iodate in buffer A (pH 4.5). Solution 3 was 5 mg/mL in buffer B (0.05 M Tris-HCl; 0.1 M NaCl) with a final pH 8.0. Solution 4 was 5 mg/mL chymotrypsin in buffer B (pH 8.0).
The bacterial cells were incubated with each of the four solutions (numbered from 1 to 4) for 1 h at 37°C. After that, the bacteria were centrifuged at 15 000 g at 0°C for 10 min, washed twice in 1 mL of PBS and resuspended in 4 mL of complete DMEM. 1 mL of each re-suspension was added to one well containing cell culture. Biological triplicates of each condition were done.
Blocking of specific carbohydrate ligands on cell wall: 3 mL of bacterial cells and broth culture supernatant were separated by centrifugation at 15 000 g at 0°C for 10 min. The bacterial cells were then washed once in PBS and then suspended in 3 mL of one of the following solutions: 15 &mgr;g/mL Concanavalin A (ConA) in PBS, 15 &mgr;g/mL succinyl Concanavalin A (succinyl-ConA) in PBS and 15 &mgr;g/mL Tetragonolobus purpurea in PBS. The cells were incubated for 30 min at 37°C, then centrifuged at 15 000 g at 0°C for 10 min, washed twice in 1.0 mL of PBS, and resuspended in 3.0 mL of complete DMEM. Treatments were done as described in the section “infection of intestinal cells”.
UV killed bacteria: To kill the bacteria, 3 mL of bacterial suspensions were exposed to UV light for 5 min[19] to make sure at least 99% of the bacteria were killed. The killed bacteria were plated on their respective agar plates to ensure cell death. After killing the bacteria, treatments were performed as described in the section “infection of intestinal cells”.
Monolayers of HCT116 were prepared by inoculating 2-chamber slides (Lab-Tek Chamber Slide, Nunc, Inc., USA) with 1.4 × 105 cells per chamber in 2 mL of complete DMEM 24 h before the adhesion assay. After 24 h, the monolayers were washed twice in 1 mL of sterile PBS. Bacterial strains were prepared and 2 mL of each resuspension was added to a chamber of the slide and incubated at 37°C with 5% CO2 for 1 h. After incubation, the monolayers were washed twice with sterile PBS then Gram stained and examined microscopically. The numbers of adherent bacteria per 100 HCT116 cells were counted in 10 random microscope areas.
Cytokines in cell culture supernatants were assayed by Enzyme Linked Immunosorbent Assays (ELISAs). After the infection (described in the sections “infection of intestinal cells” and “determination of effecter molecules”), supernatants were harvested and centrifuged for 10 min at 12 000 g to remove the bacteria. IL-8, IL-4 and IL-10 measurements were performed as the instructions in the ELISAs kits (BD bioscience, San Diego, CA). Individual samples were tested in triplicate and the concentration of cytokines was determined using the standard provided by the manufacture.
For TGF-β assays, 100 &mgr;L supernatants were transiently acidified with 2 &mgr;L 1 M HCL for 1 h at 4°C, and then neutralized with 2 &mgr;L 1 mol/L NaOH. The supernatants were then applied to TGF-β ELISAs (BD bioscience, San Diego, CA) according to the manufacturer’s instructions.
Data are expressed as means ± SD. Significance of differences was determined using the unpaired Student’s T-test and analysis of variance. P values < 0.05 were considered to be statistically significant.
A total of 27 strains of LAB were isolated from feces of new born healthy infants aged from 3 d and 1 mo on MRS agar or Slanetz/Bartley media. 29 strains of other LAB were obtained from our collection for the comparison of their modulation of inflammation cytokine (Table 1).
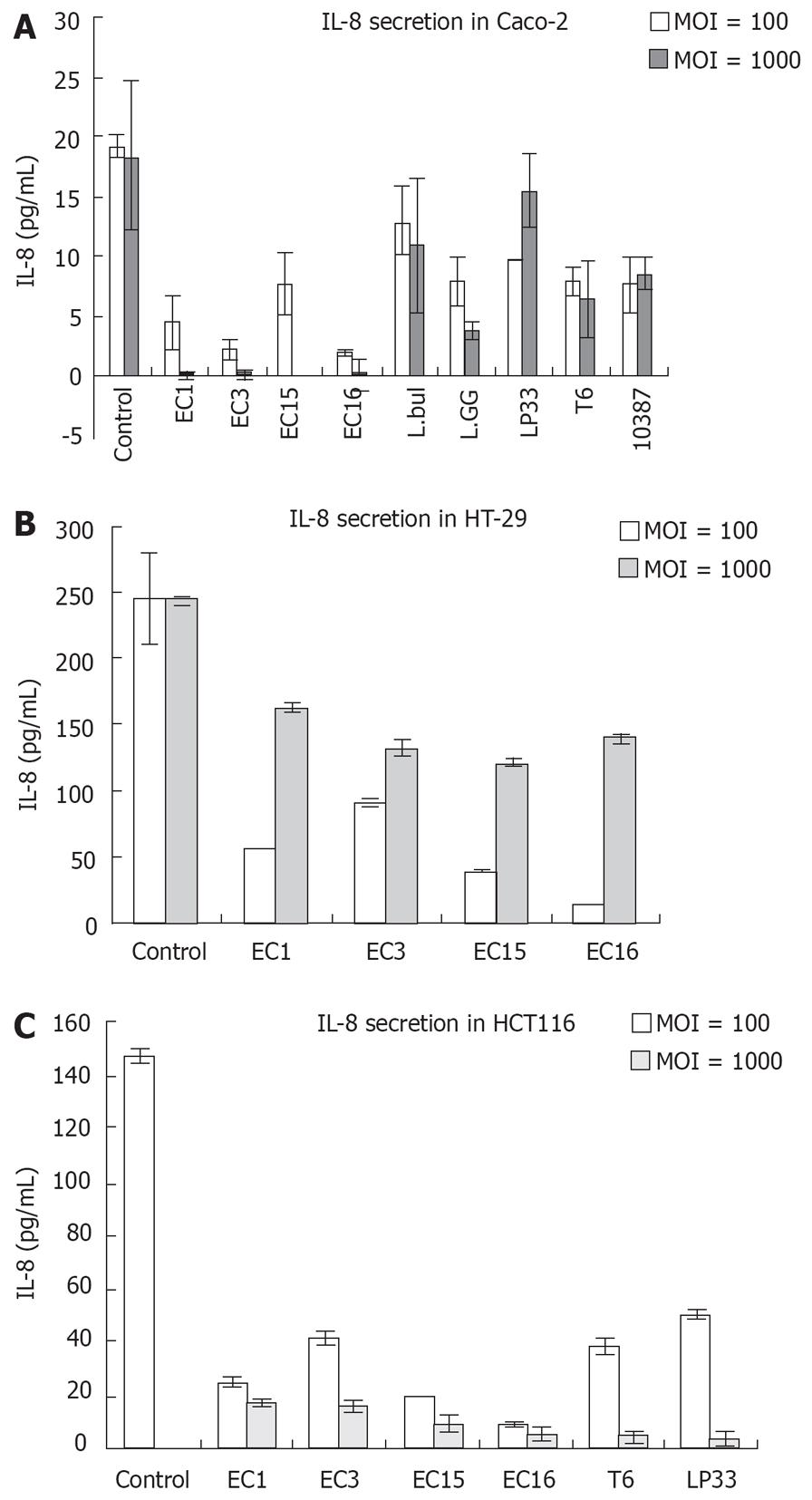
To assess the effects of LAB on IL-8 secretion in intestinal cell lines, Caco-2, HT-29 and HCT116 cells were incubated with the respective LAB strains listed in Table 1. The experiments were performed 2-5 times in replicates (Figure 1).
In our study, Caco-2, HT-29 and HCT116 continuously expressed IL-8 at a detectable level. Most of the LAB tested (including strains of E. faecalis) showed no significant regulation of IL-8 secretion (data not shown) in these three IECs. Bifidobacterium, which was reported to down-regulate TNF-α and IL-8 productions and inhibit NF-κB activation in inflamed mucosa of active ulcerative colitis[20], showed no effect on IL-8 secretion in all the three IECs tested. However, E. faecalis (EC1, EC3, EC15, EC16), L. delbrueckii bulgaricus D1 (L.bul), L. rhamnosus GG (L.GG), L. paracasei LP33 (LP33), L. brevis T6 (T6) and Streptococcus thermophilus NCIMB10387 (10387) downregulated IL-8 secretion in Caco-2 cells (Figure 1). In HT-29 cells, only E. faecalis (EC1, EC3, EC15 and EC16) could downregulate IL-8 secretion. Whereas in HCT116, E. faecalis (EC1, EC3, EC15, EC16), T6 and LP33 suppressed IL-8 secretion. Strikingly, among the 56 LAB that we have tested, E. faecalis (EC1, EC3, EC15 and EC16), which were isolated from new born healthy infants, showed suppression of IL-8 secretion in all the three IECs.
The balance between inflammatory and regulatory cytokines is of crucial importance in the gut immune modulation. TGF-β is an important inflammation regulatory cytokine. In our study, TGF-β cytokine were greatly induced by E. faecalis (EC3, EC15) (107 cfu/mL) in Caco-2 cells (data not shown). Other LAB, however, could not induce TGF-β cytokine secretion above the detection level (above 62.5 pg/mL) in IECs. IL-4 and IL-10 secretion were not above the detection level in our study (Data not shown).
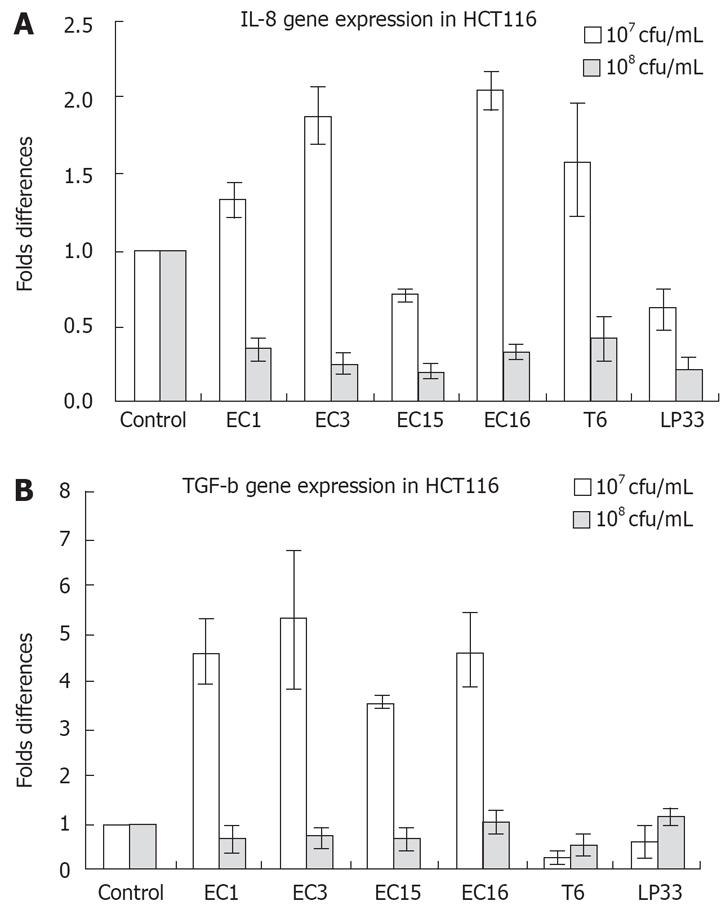
Not consistent with IL-8 secretion, addition of E. faecalis (EC1, EC3, EC15, EC16) and LP33 (107 cfu/mL) to HCT116 cells enhanced IL-8 mRNA expression (Figure 2A). However, 108 cfu/mL E. faecalis (EC1, EC3, EC16), LP33 and T6 strongly suppressed IL-8 mRNA expression in HCT116 cells (Figure 2A). Interestingly, both concentrations of LAB suppressed IL-8 secretion in HCT116 cells (Figure 1C). Almost the same was observed in Caco-2 cells: IL-8 mRNA was upregulated by 107 cfu/mL E. faecalis (EC1, EC3, EC15, EC16), L.GG and L.bul. However, there was no significant change in IL-8 mRNA expression with 108 cfu/mL LAB stimulation (Data not shown). As shown in Figure 2B, TGF-β gene expression was greatly enhanced by 107 cfu/mL E. faecalis (EC1, EC3, EC15, EC16) and T6 in HCT116 cells, however, 108 cfu/mL of LAB showed no regulation of TGF-β gene expressions.
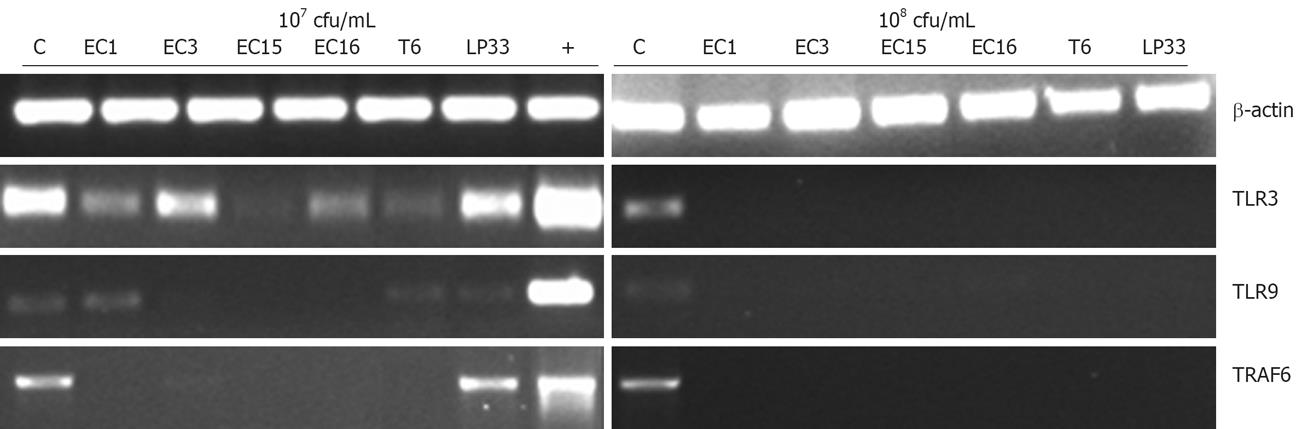
Activation of signal transduction pathway by TLRs led to the induction of inflammatory cytokine and chemokine genes that function in host defenses and tumor progression. The ability of the intestinal cell lines in response to E. faecalis and other LAB at the level of TLRs mRNA expression was therefore assessed using semi-quantitative RT-PCR. Gene encoding TLR1, TLR2, TLR3, TLR4, TLR5, TLR9, TRAF6 and Tollip were analyzed.
Examination of the TLR9 gene expression showed that TLR9 mRNA expression was greatly inhibited by 108 cfu/mL E. faecalis (EC1, EC3, EC15, EC16), T6 and LP33 treatment in HCT116 cells (Figure 3). As shown in Figure 3, TLR3 mRNA expression was also greatly inhibited by 108 cfu/mL E. faecalis (EC1, EC15, EC16), T6 and LP33 in HCT116 cells. Most interestingly, TRAF6 gene expression in HCT116 cells was totally inhibited after treatment with 107 cfu/mL (except L.bul) and 108 cfu/mL LAB (Figure 3). In HT-29 cells, E. faecalis EC1, EC3 and EC15 but not EC16 suppressed TRAF6 mRNA expression (Data not shown). This observation parallels the TLR4 regulation in HT-29 cells. TLR4 is an essential receptor that transduces the signals of bacterial cell wall lipopolysaccharide (LPS). In the TLRs signaling pathways, TLR4 could activate numerous cytokine genes via TRAF6. TLR4 was not regulated significantly by LAB in Caco-2 cells. TLRs expression in these three cell lines were summarized in Table 3.
| TLR1 | TLR3 | TLR4 | TLR5 | TLR9 | TRAF6 | TOLLIP | |
| HCT116 | NE | S | NE | NE | S | S | NC |
| Caco-2 | NE | NC | NC | NE | S | NC | NC |
| HT-29 | NE | NC | S | NE | NC | S | NC |
Since some LAB tested, especially E. faecalis, inhibited IL-8 secretion in colon cancer cell lines, we further explored the possible effecter molecules that may play a role in the anti-inflammatory effects. For simplicity, we chose E. faecalis (EC1, EC3, EC15 and EC16), L. rhamnosus GG (L.GG) and S. typhimurium (Salm) for this study. The components from the bacteria were co-cultured with HCT116 cells. IL-8 level was measured using ELISA method.
First we tested the effects of sonicated bacteria and their cell wall components on IL-8 secretion. Cell wall components from all four E. faecalis strains demonstrated similar abilities as whole cells in reducing IL-8 levels (Data not shown). Cell debris of S. typhimurium, showed the expected ability in upregulating IL-8 levels. Co-culturing with L. rhamnosus GG cell debris caused no change in IL-8 concentration. To study whether attenuation of IL-8 level required bacterial cell contents, crude bacterial cell extract was co-cultured with HCT116 cells. In contrast to the results obtained using the cell debris, the crude cell extract of all four E. faecalis strains showed no ability in reducing IL-8 level (Data not shown).
Second, we tested the need for the four E. faecalis strains to be viable to attenuate IL-8 levels. The UV killed E. faecalis maintained its ability to suppress IL-8 levels (data not shown). The data implies that inhibition of IL-8 secretion from HCT116 cells does not need E. faecalis metabolites.
Third, to further confirm whether reduction in IL-8 levels required the direct contact between HCT116 cells and bacteria or could be mediated by a secreted product from either the bacteria or the intestinal epithelial cells, conditional media was made and added to HCT116 cells. In the first treatment (Section “Infection of intestinal cells”), supernatants of the controls containing IL-8 were harvested and used in the subsequent treatment as conditional media. It was shown that E. faecalis could not reduce the IL-8 level in the conditional media containing IL-8 (data not shown). Furthermore, intestinal epithelial cell cultures were incubated with bacteria housed in sterile plastic inserts, to ensure physical separation between the HCT116 cells and bacteria while allowing secreted bacterial products to reach the HCT116 cells. The results showed that all the bacterial strains could not suppress IL-8 levels as well (data not shown). Moreover, we investigated the possible ability of the four E. faecalis strains to stimulate a release of anti-inflammatory factors by HCT116 cells as described in the section “Conditional medium and cell inserts treatment”, it was demonstrated that the supernatant harvested from co-culturing bacterial cells with HCT116 were incapable of reducing IL-8 levels in HCT116 cells.
Combining all the results, there is indication that the presence of bacterial cell wall components and attachment of bacteria to cells are essential in IL-8 attenuation.
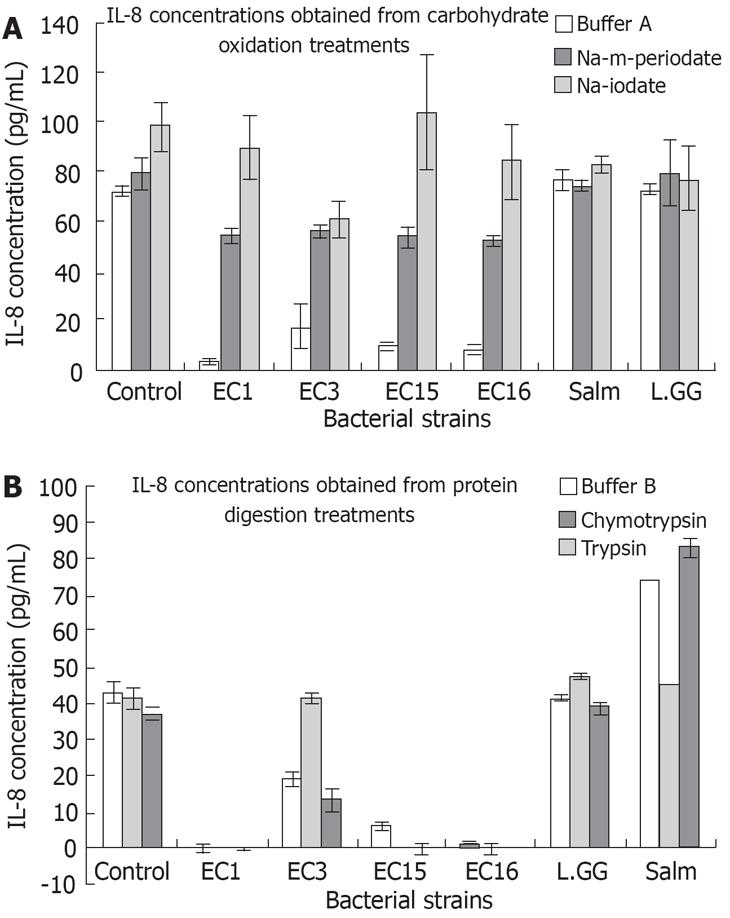
To further figure out the possible molecules on the bacterial cell wall that played the key role in the attenuation of IL-8 secretion. The carbohydrates on bacterial cell walls were oxidized using m-periodate and iodate. Results of the four E. faecalis strains in buffer A showed an expected large decrease in IL-8 levels as seen in Figure 4A. Interestingly, the Na-iodate and Na-m-periodate treatments of them prevented an appreciable decrease in IL-8 levels (Figure 4A). Results from chymotrypsin and trypsin digested bacterial cell wall proteins in buffer B showing that buffer B, chymotrypsin and trypsin did not change the abilities of the E. faecalis strains in attenuating IL-8 level (Figure 4B). However, chymotrypsin treated E. faecalis EC3 seemed unable to significantly reduce IL-8 levels as seen in Figure 4B. S. typhimurium treated with buffer B and with chymotrypsin in buffer B showed no change in its capacity to increase IL-8 levels. Surprisingly, S. typhimurium treated with trypsin in buffer B showed a heightened ability in increasing IL-8 concentrations. These results suggest that carbohydrates on the bacterial cell wall are the major effecter molecules in the regulation of IL-8 secretion. However, proteins could also be involved.
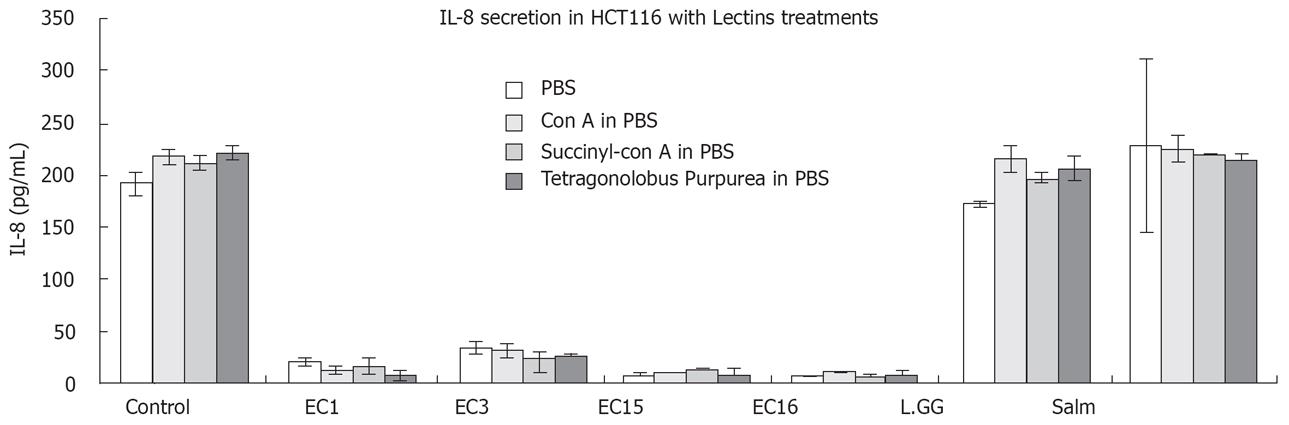
In order to test which specific carbohydrates are important in reducing IL-8 levels, bacterial strains were treated with three different lectins. Succinyl-ConA and ConA have affinity for terminal α-D-glucosyl and α-D-mannosyl residues[21]. Tetragonolobus purpureas (TPA) on the other hand as an affinity for α-L-fucosyl residues and an especially high affinity for α-L-fucose residues on type II chain blood group oligosaccharides[22]. These lectins would bind specifically to these residues and possibly change the overall carbohydrate structures, potentially affecting their role in attenuating IL-8 secretion. Figure 5 showed that the three lectins used also had no effect on depressing IL-8 secretion. Interestingly, with the employment of lectins, the ability to activate IL-8 by S. typhimurium was inhibited. This might imply that lectins may act on the carbohydrate in S. typhimurium to eliminate its stimulation on IL-8 secretion.
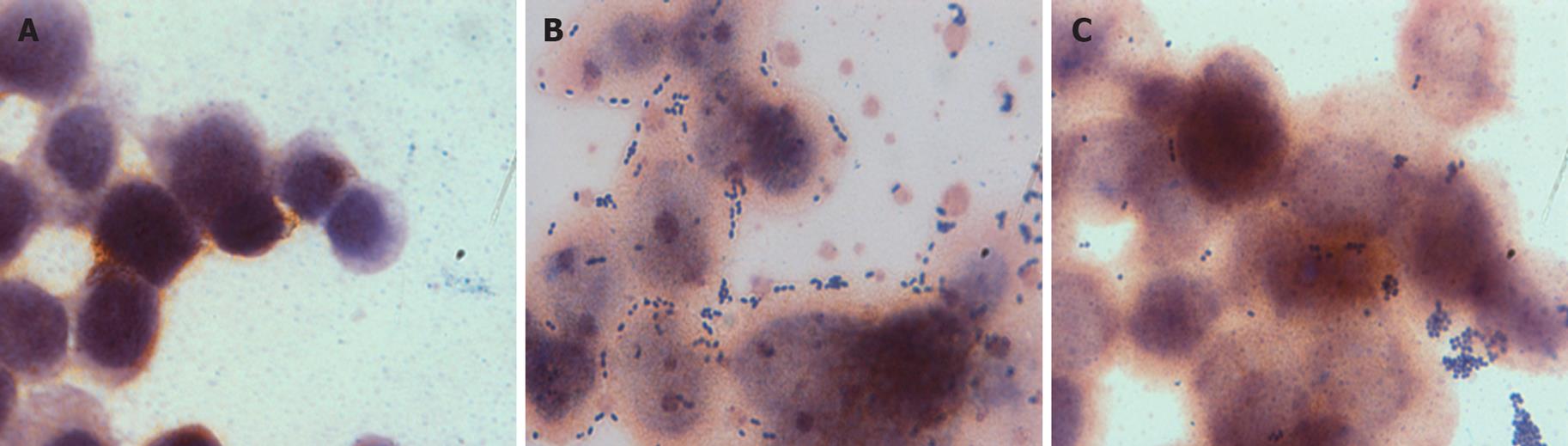
In order to study the importance of cell wall carbohydrates in adhering to HCT116 cells, carbohydrates on bacterial cell surfaces were oxidized using m-periodate. Figure 6 demonstrates that m-periodate treatment significantly reduced adhesion of all four E. faecalis strains to HCT116 cells (Table 4). Moreover, the secretion of IL-8 in HCT116 was greatly upregulated (Figure 4A). This study implies that carbohydrates on the bacterial cell wall helped bacteria to adhere to cells and the carbohydrate could act as ligands to activate signaling pathways to suppress IL-8 secretion from IECs. The carbohydrate could be structure component since sonicated and UV killed E. faecalis strains were able to suppress IL-8 secretion.
In the present study, we investigate the immune modulatory effects of infant intestinal E. faecalis compared with some other LABs on intestinal cells. Human intestinal cells (Caco-2, HT-29 and HCT116) have been shown to constitutively produce IL-8 in our study. IL-8, which is a potent neutrophil chemo-attractant and plays a key role in allergic inflammation, may be involved in the E. faecalis regulation of host immunology. The suppression of IL-8 secretion showed the reduction of inflammation reactions in animal and clinical trials[23]. In this study, most of the LABs tested, including most strains of E. faecalis (12 out of 16 strains isolated), did not show any effect on IL-8 production. However, IL-8 secretion was greatly downregulated after exposure to E. faecalis strains EC1, EC3, EC15, EC16 and some other LAB in IECs. This suggests that the immune reactions of an individual could be modulated by specific strains of a bacterial species, and his well being is determined by the specific strains of LAB which were established during early childhood. The inhibition of IL-8 protein secretion by E. faecalis could explain the increased incidence of allergy in infants deprived of E. faecalis in the intestine[68]. Although all 4 strains of E. faecalis and other LABs could inhibit the secretion of IL-8 (Figure 1), it is interesting to note that the different bacteria were reduced to a different level of IL-8 secretion after their exposure. E. faecalis in general suppressed IL-8 production to a higher degree than the other LABs. The downregulation of IL-8 secretion is dose dependent in Caco-2 and HCT116 cells. The higher the concentration of LAB, the less secretion of IL-8. Paradoxically, some of the IL-8 secretion was not parallel with its mRNA expression in the intestinal cell lines in response to the LAB. IL-8 mRNA was enhanced; however, its protein secretion was inhibited in IECs when they were exposed to E. faecalis at a concentration of 107 cfu/mL. More interestingly, IL-8 protein secretion was enhanced however its mRNA expression remained unchanged after the challenge of S. typhimurium at a concentration of 107 cfu/mL (data not shown). This implies that E. faecalis could boost the immune system to be ready for the challenge of pathogen at a physiological concentration (107 cfu/mL). However, the inflammatory responses were suppressed in the absence of pathogen and the presence of E. faecalis. On the contrary, S. typhimurium activated inflammatory responses without upregulating the immune gene expression. Thus, the regulation of IL-8 is important in the immune homeostasis. Its modulation by E. faecalis could involve post-transcriptional processes in protein synthesis and secretion[24]. The suppression of IL-8 at colon cancer cell lines’ basal level could inhibit its inflammatory responses. Furthermore, these data suggests that IECs could differentiate commensal bacteria from pathogenic bacteria and thus lead to different immune responses.
Interestingly, TGF-β, which is important in inflamma-tory disease recovery, was upregulated significantly by E. faecalis EC3 and EC15 in Caco-2 cells at protein level and in HCT116 at the gene level. TGF-β is also a key factor implicated in the regulation of intestinal barrier function and is thought to mediate tolerance to the indigenous microflora. Compared with Lactobacillus and Bifidobacterium, E. faecalis demonstrated stronger effects on TGF-β production. The suppression of IL-8 secretion and activation of TGF-β production in the presence of E. faecalis suggested that E. faecalis could alter the balance among Th1 and Th2 immunities and leading to prevention of inflammation in intestinal cells.
Toll like receptors (TLRs) are also important in regulating inflammatory responses and Th1/Th2 immunities in the host. In our study, TLR3, TLR4, TLR9 were found to express in some of the colon cancer cell lines and their expression was suppressed by E. faecalis. Moreover, we found that TRAF6 mRNA was also suppressed by E. faecalis and some of the LABs in IECs. TRAF6 is an adaptor in TLRs signaling pathway. It could bind TAB1, TAK1 and TAB2 to form a complex leading to the activation of NF-κB and MAP kinases[25]. Thus, the suppression of TRAF6 could inhibit the activation of the NF-κB and the MAPKs families and further the inflammatory responses. The inhibition of TLR3 and TLR9 signaling by LAB may explain the suppression of pro-inflammatory cytokines secretion in the colon cell lines. Hence, the suppression of TLR3 and TLR9 signaling could be used for therapeutic treatment of inflammatory diseases including cancer, asthma and allergy[2627]. In addition, the low expression of TLR4 in IECs may explain the tolerance of the large quantities of luminal LPS in the intestine[28]. Furthermore, recent studies showed that bacteria-induced experimental colitis involves the activation of TLR[29]. The blocking of TLRs by E. faecalis may represent an attractive strategy to treat intestinal inflammation.
Overall, the modulation of intestinal immunity by E. faecalis may involve TRAF6, a number of TLRs (TLR3, TLR4, and TLR9) and cytokines (IL-8, TGF-β).
In order to characterize the effecter molecules that modulate inflammatory responses in IECs, studies were further carried out using conditional medium. The use of conditional medium showed that the molecules were not secreted by cells or bacteria or molecules activated after co-culturing the cells and bacteria. This finding was further confirmed using cell inserts. No IL-8 downregulation was observed when the cells and bacteria were separated by cell inserts. However, when bacterial cell wall fragments were applied to the intestinal cells after sonication, there was a dramatic suppression of IL-8 secretion in HCT116. In addition, the remaining supernatant, the crude cell extract containing bacterial cytoplasm, ribosomes, small membrane vesicles, and large membrane structures could not suppress IL-8 secretion. Therefore, the molecules are not inside the bacteria but on the surface of bacterial cell walls. Since only bacterial cell walls could inhibit IL-8 secretion, it implies that the inhibition does not involve bacterial metabolites. Another study using UV killed bacteria confirmed that those killed bacteria retained the ability to suppress IL-8 secretion in HCT116.
Results from oxidation of carbohydrate moieties on the whole cell surfaces of E. faecalis showed that the four E. faecalis lost their ability to depress IL-8 secretion. Thus, carbohydrates are the main effecter molecules on E. faecalis. This mild periodate oxidation at acid pH cleaves carbohydrate vicinal hydroxyl groups without changing polypeptide chain structures[30]. Periodate consumption and iodate formation, give insight to the number of vicinal dihydroxyl groups which possibly play the roles in suppression of inflammation[31]. Chymotrypsin-mediated protein digestion of the effecter molecules, on E. faecalis EC3’s cell surface, stripped it of its ability to attenuate IL-8 levels. Thus E. faecalis EC3’s effecter molecule probably has protein structure(s) necessary for depressing IL-8 secretion. Similar chymotrypsin-mediated protein digestion of the three other E. faecalis strains and trypsin-mediated protein digestion on all four E. faecalis strains failed to alter their ability in reducing IL-8 levels, suggesting that protein might not be the major components from the bacterial cell wall in suppressing IL-8 secretion.
The specific carbohydrate monosaccharides involved in IL-8 level attenuation could not be determined from the lectin study in our experiments. This study suggests that certain monosaccharides that are not part of the inflammation suppressing effecter molecule. α-L-fucosyl, α-L-fucose, D-mannosyl and D-glucosyl residues are probably not vital or possibly absent from the effecter molecule. However they might be important in upregulating IL-8 secretion in S. typhimurium.
The adhesion study of E. faecalis strains demonstrated that they adhered well to the mucosal layer of epithelial cells. Oxidation of the carbohydrate moieties on the bacterial cell surface caused a decrease in their adhesive abilities as well as a drop in the ability to suppress IL-8 secretion. This suggests that the cell surface carbohydrates are necessary for the adhesion of four E. faecalis strains to IECs. This is in congruence with Guzmàn et al who previously observed that enterococcal surface carbohydrate adhesins were necessary for binding of E. faecalis to Girardi heart human cells and urinary tract epithelial cells[32]. Other studies by Shorrock and Lambert also demonstrated that carbohydrate oxidation reduced the binding abilities of E. faecalis, suggesting that surface adhesin with the carbohydrate component is necessary for adhesion[33]. Since the same oxidation of carbohydrate moieties also stripped these enterococci of their IL-8 secretion suppressing abilities, adhesion of these bacteria via carbohydrate moieties plays a vital role in suppressing inflammation.
In conclusion, the study has demonstrated that E. faecalis is the major human intestinal LAB in modulating expression of host immune genes that participate in inflammation. Moreover, the immune regulatory mechanism may involve the adhesion of E. faecalis on intestinal cells and suppression of specific TLRs signaling pathways via carbohydrate mediated adhesion-receptor interaction.
It has long been assumed that indigenous intestinal microbiotas are involved in the immunomodulation of the host and the effects are strain dependent. However, there are few direct evidences in support of the assumption. In addition, the underlying molecular and cellular mechanisms are unclear.
Recent studies showed that Enterococcus faecalis could suppress the proliferation of intestinal pathogens and thus may prevent infection and induction of inflammation. Studies on Bacteroides thetaiotaomicron, a prevalent commensal bacterium of the human intestine showed that it might attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and Rel A. Toll like receptors (TLRs) and cytokines are involved in the anti-inflammation process.
In our knowledge, this is the first time E.faecalis is demonstrated as the main immune modulator in the intestinal Lactic acid bacteria (LAB) of infants, and cell wall carbohydrate is involved in the regulatory effects.
This finding highlights possible new approaches for therapeutic treatment of inflammatory disease in humans. The study suggests that the immunological status in the gastrointestinal tract of infants may be determined by the presence of specific strains of E. faecalis.
Probiotics: Live microorganisms which can when administered in adequate amounts confer a health benefit on the host; LAB: Lactic acid bacteria comprise a clade of Gram positive, low-GC, acid tolerant, non-sporulating, non-respiring rod or cocci that are associated by their common metabolic and physiological characteristics. LAB is the most common type of microbe used as a probiotic.
Wang et al showed that some Enterococcus strains reduced IL-8 secretion from intestinal epithelial cells. Focus of this manuscript is interesting.
| 1. | Cross ML, Gill HS. Can immunoregulatory lactic acid bacteria be used as dietary supplements to limit allergies? Int Arch Allergy Immunol. 2001;125:112-119. |
| 2. | Marteau PR. Probiotics in clinical conditions. Clin Rev Allergy Immunol. 2002;22:255-273. |
| 3. | Abreu MT, Vora P, Faure E, Thomas LS, Arnold ET, Arditi M. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J Immunol. 2001;167:1609-1616. |
| 4. | Lan JG, Cruickshank SM, Singh JC, Farrar M, Lodge JP, Felsburg PJ, Carding SR. Different cytokine response of primary colonic epithelial cells to commensal bacteria. World J Gastroenterol. 2005;11:3375-3384. |
| 5. | Janeway CA, Travers P, Walport M, Shlomchik M. Immunobiology: the immune system in health and disease. 5th ed. New York: Garland Publishing 2001; 408. |
| 6. | Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516-520. |
| 7. | Björkstén B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy. 1999;29:342-346. |
| 8. | Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut. 2002;51:51-55. |
| 9. | Lammers KM, Helwig U, Swennen E, Rizzello F, Venturi A, Caramelli E, Kamm MA, Brigidi P, Gionchetti P, Campieri M. Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol. 2002;97:1182-1186. |
| 11. | Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-beta/Smad signaling and fail to inhibit proinflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J Immunol. 2005;174:2990-2999. |
| 12. | McKay AM. Antimicrobial activity of Enterococcus faecium against Listeria spp. Lett Appl Microbiol. 1990;11:15-17. |
| 13. | Giraffa G. Enterococcal bacteriocins: their potential as anti-Listeria factors in dairy technology. Food Microbiol. 1995;12:291-299. |
| 14. | Oh DH, Marshall DL. Antimicrobial activity of ethanol, glycerol monolaurate or lactic acid against Listeria monocytogenes. Int J Food Microbiol. 1993;20:239-246. |
| 15. | Isolauri E, Kirjavainen PV, Salminen S. Probiotics: a role in the treatment of intestinal infection and inflammation? Gut. 2002;50 Suppl 3:III54-III59. |
| 16. | Shimada T, Cheng L, Enomoto T, Yang X, Miyoshi A, Shirakawa T. Lysed enterococcus faecalis FK-23 oral administration reveals inverse association between tuberculin responses and clinical manifestations in perennial allergic rhinitis: a pilot study. J Investig Allergol Clin Immunol. 2004;14(3):187-192. |
| 17. | Amano J, Oshima M. Expression of the H type 1 blood group antigen during enterocytic differentiation of Caco-2 cells. J Biol Chem. 1999;274:21209-21216. |
| 18. | Shon W, Lim S, Bae KS, Baek S, Lee W. The expression of alpha4 integrins by human polymorphonuclear neutrophils in response to sonicated extracts of Enterococcus faecalis. J Endod. 2005;31:369-372. |
| 19. | Conner-Kerr TA, Sullivan PK, Gaillard J, Franklin ME, Jones RM. The effects of ultraviolet radiation on antibiotic-resistant bacteria in vitro. Ostomy Wound Manage. 1998;44:50-56. |
| 20. | Bai AP, Ouyang Q, Xiao XR, Li SF. Probiotics modulate inflammatory cytokine secretion from inflamed mucosa in active ulcerative colitis. Int J Clin Pract. 2006;60:284-288. |
| 21. | Coniglio L, Morale A, Angelini C, Falugi C. Cholinergic activation of settlement in Ciona intestinalis metamorphosing larvae. J Exp Zool. 1998;280:314-320. |
| 22. | Seddas P, Boissinot S. Glycosylation of beet western yellows virus proteins is implicated in the aphid transmission of the virus. Arch Virol. 2006;151:967-984. |
| 23. | MacDermott RP. Alterations of the mucosal immune system in inflammatory bowel disease. J Gastroenterol. 1996;31:907-916. |
| 24. | Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72:847-855. |
| 25. | Akira S, Sato S. Toll-like receptors and their signaling mechanisms. Scand J Infect Dis. 2003;35:555-562. |
| 26. | Salaun B, Coste I, Rissoan MC, Lebecque SJ, Renno T. TLR3 can directly trigger apoptosis in human cancer cells. J Immunol. 2006;176:4894-4901. |
| 27. | Bhattacharjee RN, Akira S. Modifying toll-like receptor 9 signaling for therapeutic use. Mini Rev Med Chem. 2006;6:287-291. |
| 28. | Nomura F, Akashi S, Sakao Y, Sato S, Kawai T, Matsumoto M, Nakanishi K, Kimoto M, Miyake K, Takeda K. Cutting edge: endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface toll-like receptor 4 expression. J Immunol. 2000;164:3476-3479. |
| 29. | Karrasch T, Kim JS, Muhlbauer M, Magness ST, Jobin C. Gnotobiotic IL-10-/-;NF-kappa B(EGFP) mice reveal the critical role of TLR/NF-kappa B signaling in commensal bacteria-induced colitis. J Immunol. 2007;178:6522-6532. |
| 30. | BOBBITT JM. Periodate oxidation of carbohydrates. Adv Carbohydr Chem. 1956;48:1-41. |
| 31. | Honda S, Suzuki K, Kakehi K. Simultaneous determination of iodate and periodate by capillary zone electrophoresis: application to carbohydrate analysis. Anal Biochem. 1989;177:62-66. |
| 32. | Guzmàn CA, Pruzzo C, Platè M, Guardati MC, Calegari L. Serum dependent expression of Enterococcus faecalis adhesins involved in the colonization of heart cells. Microb Pathog. 1991;11:399-409. |
| 33. | Shorrock PJ, Lambert PA. Binding of fibronectin and albumin to Enterococcus (Streptococcus) faecalis. Microb Pathog. 1989;6:61-67. |