Published online Jan 28, 2008. doi: 10.3748/wjg.14.574
Revised: October 8, 2007
Published online: January 28, 2008
AIM: To analyze the modulation of gene expression profile associated with inhibition of liver regeneration in hepatitis B X (HBx)-expressing transgenic mice.
METHODS: Microarray technology was performed on liver tissue obtained from 4 control (LacZ) and 4 transgenic mice (HBx-LacZ), 48 h after partial hepatectomy. The significance of the normalized log-ratios was assessed for each gene, using robust t-tests under an empirical Bayes approach. Microarray hybridization data was verified on selected genes by quantitative PCR.
RESULTS: The comparison of gene expression patterns showed a consistent modulation of the expression of 26 genes, most of which are implicated in liver regeneration. Up-regulated genes included DNA repair proteins (Rad-52, MSH6) and transmembrane proteins (syndecan 4, tetraspanin), while down-regulated genes were connected to the regulation of transcription (histone deacetylase, Zfp90, MyoD1) and were involved in the cholesterol metabolic pathway and isoprenoid biosynthesis (farnesyl diphosphate synthase, Cyp7b1, geranylgeranyl diphosphate synthase, SAA3).
CONCLUSION: Our results provide a novel insight into the biological activities of HBx, implicated in the inhibition of liver regeneration.
- Citation: Sidorkiewicz M, Jais JP, Tralhao G, Morosan S, Giannini C, Brezillon N, Soussan P, Delpuech O, Kremsdorf D. Gene modulation associated with inhibition of liver regeneration in hepatitis B virus X transgenic mice. World J Gastroenterol 2008; 14(4): 574-581
- URL: https://www.wjgnet.com/1007-9327/full/v14/i4/574.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.574
The liver has a unique capacity to regenerate after partial hepatectomy (PH) or injury[1]. Experimental evidence has confirmed that hepatocyte proliferation is responsible for liver regeneration following PH, despite the low replication rate of hepatocytes in a normal liver[1]. After a two-third partial hepatectomy, hepatocytes switch from a quiescent state to a proliferative state and re-enter cell cycle division to compensate for the loss of liver mass. Liver regeneration is a multi-step process comprising at least two critical points: the immediate early gene phase (priming) that mediates the transition of the quiescent hepatocytes to the cell cycle (G1), and progression from G1 to S phase. In mice, DNA synthesis peaks at 40-44 h after PH[1].
Numerous growth factors and cytokines regulate the regeneration process by providing stimulatory and inhibitory signals for proliferation. Tumour necrosis factor alpha (TNF-α) and interleukin-6 (Il-6), together with their downstream transcription factors Stat-3 and nuclear factor kappa B (NF-κB), are the most important initiating factors in the regenerative response[1]. TNF-α-induced hepatocyte proliferation has been shown to involve the small GTPases (Ras and RhoA) necessary for cell cycle progression by stimulating the degradation of Cdk inhibitors (p21Cip and p27Kip1)[2]. The initiation factors governing liver regeneration partially overlap with regulators of the hepatic acute phase response (APR), which results in dramatic changes in the expression of acute phase response proteins following PH[3]. Hepatocyte growth factor (HGF) and transforming growth factor alpha (TGFα) further stimulate cell cycle progression, DNA synthesis and cell proliferation. Finally, TGF-β and activin suppress cell growth and terminate liver regeneration at a fixed point[1]. Recently, a large scale gene expression analysis, based on cDNA microarray technology, has provided a unique opportunity to elucidate the expression pattern throughout the course of liver regeneration[4–6], and the priming phase of liver regeneration in mice[7].
In a previous report, we showed that hepatitis B virus (HBV) X protein (HBx) inhibits liver regeneration after PH in HBx-expressing transgenic mice, and also demonstrated a paracrine inhibitory effect of HBx on liver cell proliferation[8]. HBx exhibits pleiotropic effects that modulate cell responses to genotoxic stress, protein degradation, cell viability and signalling pathways[9]. HBx has been shown to enhance transcription in the cytoplasm by activating various signal transduction cascades such as Ras/Raf mitogen-activated protein kinase and Src kinases, as well as in the nucleus through transcription factors like AP-1, AP-2 and NF-κB. Several studies have identified possible cellular targets of HBx, including members of the cyclic-AMP response element binding protein (CREB)/activating transcription factor (ATF) family, the TATA-binding protein, RNA polymerase subunit RPB5, the UV-damaged DNA-binding protein and the replicative senescence p55sen[9,10]. HBx has been shown to interact with p53 and inhibit its function, but it may also induce apoptosis via p53-dependent and independent mechanisms[9]. Furthermore, it has been demonstrated that mutations identified in HBx-encoding sequences in the tumour cells of HCC can markedly modify the biological activity of HBx, and possibly favour cell transformation[11]. It has been shown that the HBx protein participates in the development of hepatocellular carcinoma (HCC) in HBV infected patients[12]. Indeed, HBx protein can induce HCC in certain transgenic mice and may, under certain conditions, cooperate in vitro with Myc or Ras oncogene[13]. However, the mechanisms involved in HBx-mediated HCC remain obscure.
In the present study, we analyzed further the impact of HBx on gene expression profile in the liver of transgenic mice, following PH. For this purpose, we employed HBx-expressing transgenic mice mentioned above, and adopted a transcriptome approach to analyse gene expression modulation, 48 h after PH. We observed that impaired DNA synthesis and reduction in liver mass in HBx-transgenic mice was associated with a weaker expression of genes involved in the cholesterol and nonsterol isoprenoid pathways as well as in impaired expression of serum amyloid A.
Two groups of mice (the same as described previously[8]) were used as liver RNA source. HNF1-LacZ transgenic mice carrying the nls-LacZ gene driven by the hepatocyte nuclear factor (HNF-1) promoter acted as controls. Test animals AX16/HNF1-LacZ double transgenic mice, were generated by crossing homozygous AX16 transgenic mice carrying ORF HBx linked to the promoter/enhancer of human anti-thrombin III gene, with HNF1-LacZ transgenic mice. PH was performed on 21-35 d old LacZ and HBx-LacZ (with confirmed expression of HBx) transgenic mice. Frozen liver tissue obtained from 4 control (LacZ) and 4 treated (HBx-LacZ) mice was pulverized in liquid nitrogen, 48 h after partial hepatectomy. Total RNA extraction was performed using RNAble (Eurobio), according to the manufacturer’s instructions and purified by RNeasy kit (Qiagen) followed by on-column DNase digestion (RNase-free DNase set). Only RNAs with an absorbance ratio of 260/280 over 1.9 and intact ribosomal 28S and 18S rRNA bands, evaluated by 1% agarose gel electrophoresis, were utilized for the microarray study.
The aminoallyl indirect labelling method was used to obtain Cyanin-3 (Cy-3) or Cy-5 labelled cDNA. Briefly, 20 &mgr;g of total RNA and 5 &mgr;g of oligo-dT primers were incubated at 70°C for 10 min and snap-cooled in ice, in order to generate aminoallyl-labelled cDNA. Reverse transcription was then performed by adding 0.5 mmol/L dATP, dCTP, dGTP, 0.3 mmol/L dTTP, 0.2 mmol/L aminoallyl-dUTP (aa-dUTP), 400 U SuperScript II, 10 mmol/L DTT, and 1 × first strand buffer. This mixture was incubated at 42°C. After RNA hydrolysis (1 U of RNaseH at 37°C for 15 min), unincorporated aa-dUTP was removed by the QIAquick PCR purification kit (Qiagen). The resulting aminoallyl -cDNAs were diluted in 0.1 mol/L sodium carbonate (pH 9.0) and coupled with 100 &mgr;g of Cy-3 or Cy-5 monoreactive dye (Amersham), prepared in dimethyl sulfoxide (DMSO) for 1 h. Cy3 and Cy5 labelled cDNA targets were concentrated in a Microcon filter device (Millipore) and suspended in 15 &mgr;L of hybridization buffer (0.7% SDS, 70% formamide in 3.6 × Denhardt solution). The cDNA microarray consisting of 5376 mice and rat cDNA spotted onto glass slides (Functional Genomic Service, CEA) was prehybridized in buffer containing: 3.5 × sodium chloride-sodium citrate buffer (SSC), 1% Bovine Serum Albumin (BSA) and 0.1% sodium dodecyl sulfate (SDS) for 1 h at 50°C. For each experiment, test cDNA was mixed with control cDNA, labelled with Cy3. Mixed, labelled cDNA, after preheating to 42°C in hybridization buffer, was overlaid on dried glass cDNA array and incubated overnight at 42°C. After hybridization, the slides were washed once in 2 × SSC/0.2% SDS buffer and twice in 0.2 × SSC buffer, and dried by centrifugation.
For each slide, the fluorescent images of hybridized microarray were scanned with a Gene Pix 4000B scanner. Image analysis was performed using a Gene Pix Pro 40 054 to quantify the arrays. Spots flagged automatically by Genepix or by visual inspection of the array scans were excluded from the analysis. Global normalization was applied to correct the artefacts caused by different incorporation rates or scanner settings for two dyes. Scatter plots in a log scale were performed to visualize fold changes between two channels by plotting Cy5 intensity against Cy3 intensity. Normalization steps were performed using Bioconductor array packages[14]. After removal of the background, we normalized the log2 of the cy5/cy3 ratio using a print-tip loess approach[15]. For each gene, the average of the normalized log-ratios was computed and the significance was assessed using robust t-tests under an empirical Bayes approach[16] as implemented in the limma package. P-values were then corrected by the FDR approach[17] to take account of test multiplicity. Differentially expressed genes were considered to be those with a false discovery rate (FDR) of less than 5% and an expression ratio > 1.4.
In order to verify microarray hybridization data, real-Time PCR was performed using a Light Cycler rapid thermal system (Roche Diagnostics). Random cDNA was transcribed from 10 &mgr;g total RNA with 400 U of Superscript II in the presence of 1 &mgr;g random primers, according to the manufacturer’s instructions. All PCR experiments were carried out on the same cDNA preparation. Reactions were performed according to the manufacturer’s instructions in a 20 &mgr;L volume containing 2 &mgr;L of 10-fold diluted cDNA, 0.5 &mgr;mol/L of primers, and an MgCl2 concentration optimized between 2 and 4 mmol/L. A typical protocol included 10 min of initial denaturation followed by 40 cycles of 95°C denaturation for 20 s, annealing at 53°C-56°C for 15 s and a 72°C extension for 5-12 s (depending on the PCR product). For each set of primers, a template without reverse transcription was amplified as a negative control. The details of primer sequences and amplification conditions are summarized in Table 1. The quantification of gene expression was based on a standard curve prepared from gene-specific, purified PCR product (PCR clean-up kit, Macherey-Nagel). The specificity of each PCR product was controlled by a melting curve analysis and subsequent agarose gel electrophoresis of the PCR product. For each experiment, relative concentrations were obtained after normalization with β-actin values. Calculation of the ratio of each mRNA expression was based on the relative concentration of specific cDNA found by RT-PCR in samples originating from control and HBx-expressing livers.
| Name of gene | Sequences of forward (F) and reverse (R) primers | Source of primers | PCR product (bp) | T anneling (°C) | Mg2+ (mmol/L) |
| Serum amyloid A3 | F: TCAGCACATTGGGATGTTTAGG | UniSTS: 219434 | 206 | 53 | 3 |
| R: CAGAGGACTCAAGAGCTGACCA | |||||
| Creatine kinase muscle | F: CCTCCTGGAAGTCCAATCAT | UniSTS: 159603 | 150 | 53 | 3 |
| R: GGCCATCACGGACTTTTATT | |||||
| Peroxiredoxin 1 | F: GAGCAGCCAGAAGAAACTCTTG | UniSTS: 144118 | 153 | 53 | 3 |
| R: AGAAGATTGGTCTGCCCAAAA | |||||
| Retinoblastoma-like 2 | F: TGGCTGAGTCCTGTAACAAC | UniSTS: 162222 | 374 | 53 | 2 |
| R: CCAACACCTTTCTGAGGC | |||||
| Interleukin-6-receptor, 80-kD | F: AAGCAGCAGGCAATGTTACC | [18] | 120 | 55 | 3 |
| R: CATAAATAGTTCCCAGTGTCG | |||||
| DNA mismatch repair protein MSH6 | F: ATATGTCCTAGGCGCACACAAA | UniSTS: 211063 | 208 | 56 | 4 |
| R: CTAGCATACTCAGGCATGCGAC | |||||
| Peroxisome proliferator-activated receptor-α | F: CATCGAGTGTCGAATATGTGG | [19] | 172 | 55 | 4 |
| R: GCAGTACTGGCATTTGTTCC | |||||
| β-actin | F: CGTGACATTAAGGAGAAGCTGTGC | [20] | 374 | 53 | 2 |
| R: CTCAGGAGGAGCAATGATCTTGAT |
As previously reported, HBx expression inhibits liver regeneration after PH in AX16/HNF1-LacZ transgenic mice, by a combination of intracellular and paracrine effects[8]. In the present study, we investigated, using microarray analysis consisting of 5376 murine genes, the impact of HBx expression on gene expression profile, 48 h after partial hepatectomy to analyze any changes in gene expression which may be involved in the observed inhibition of liver regeneration in HBx transgenic mice. This time point of gene expression analysis was chosen in order to determine alterations in gene expression associated with the DNA synthesis phase[1]. In addition, BrdU incorporation analysis performed on SCID mice transplanted with HBx-expressing hepatocytes, 48 h after partial hepatectomy, had indicated a marked (5.5-fold) reduction in the cellular DNA synthesis[8].
RNAs isolated from the livers of four HBx-LacZ mice were labelled separately with Cy5, and RNAs from the livers of four LacZ mice were mixed together and labelled with Cy3 as the reference. Thus, the expression profile of each of the four tested RNA was compared with the expression of control RNA by the microarray assay (performed in duplicate). 82% of the clones spotted on the microarray revealed hybridization signals, and the data shown in Tables 2 and 3 was obtained after the elimination of genes that did not comply with the criteria for expression, normalization and statistical analysis (see Material and Methods). In order to validate the microarray analysis, real-time PCR was performed on selected genes for which the expression was found to be up- or down-regulated or unmodified in LacZ-HBx mice when compared with LacZ mice. As shown in Table 4, the RT-PCR results were similar to the data obtained from cDNA microarrays; we therefore did not perform RT-PCR on all the genes listed in Tables 2 and 3.
| Gene name (Abbreviation) | Unigene ID | GB Acc. | Fold difference HBx-LacZ/LacZ | P value | Function |
| Aven: caspase activation inhibitor | Mm.292041 | BF662037 | 1.50 | 0.030 | Caspase inhibitor |
| RAD52 homolog | Mm.149 | U12135 | 2.14 | 0.035 | DNA repair/double-strand break |
| Mismatch repair protein MSH6 | Rn.16755 | XM345633 | 1.51 | 0.009 | DNA repair/mismatch |
| Epidermal growth factor | Rn.6075 | NM012842 | 1.40 | 0.014 | Growth factor |
| DC-SING (CD209) | Mm.32510 | NM133238 | 1.40 | 0.009 | Transmembrane protein/Cell adhesion |
| Leucyl-tRNA synthetase | Hs.432674 | NM020117 | 1.41 | 0.009 | Metabolism/protein synthesis |
| Syndecan 4 | Mm.3815 | BC005679 | 1.42 | 0.014 | Transmembrane protein/Focal adhesion |
| Transmembrane 4 superfamily member | Mm.18590 | BC050153 | 1.45 | 0.009 | Tetraspanin interacting with beta-1 integrin |
| Golgi SNAP receptor complex member 2 | Rn.13518 | BC061994 | 1.42 | 0.011 | Transmembrane protein/Vesicular transport of Golgi |
| Gene name (Abbreviation) | Unigene ID | GB Acc. | Fold difference HBx-LacZ/LacZ | P value | Function |
| Serum amyloid A3 (Saa3) | Mm.14277 | X03479 | -1.95 | 0.016 | Acute phase response protein cholesterol transport |
| Cadherin 16 (Cdh 16) | Mm.19423 | AF016271 | -2.64 | 0.010 | cell recognition protein |
| Creatine kinase (Ckm) | Mm.2375 | AI325205 | -1.65 | 0.040 | ATP synthesis |
| Glutathione S-transferase, pi2 (Gstp2) | Mm.299292 | AI325120 | -1.85 | 0.043 | Detoxification and drug metabolism |
| Progastricsin ( pepsinogen C) (Pgc) | Mm.22957 | AK008959 | -1.73 | 0.009 | Digestion enzyme |
| Phospholipase A2 group 1B (Pla2g1b) | Mm.20190 | AI327450 | -1.58 | 0.016 | Digestion enzyme |
| Tripsin II precursor (Try2) | Mm.301947 | AI386046 | -1.54 | 0.014 | Digestion enzyme |
| Farnesyl diphosphate synthase (Fdps) | Mm.39472 | W76783 | -1.98 | 0.020 | Enzyme of cholesterol pathway |
| Cytochrom P450, 7b1 (Cyp7b1) | Mm.278588 | U36993 | -1.61 | 0.009 | Enzyme of cholesterol pathway |
| Geranylgeranyl diphosphate synthase 1 (Ggps 1) | Mm.148039 | AB016044 | -1.41 | 0.034 | Enzyme of cholesterol pathway |
| Aldolase1, A isoform (Aldo 1) | Mm.275831 | AI327494 | -3.35 | 0.015 | Enzyme of glycolysis pathway |
| Peptidylglycine alpha-amidating monooxygenase | Mm.5121 | AI323455 | -1.73 | 0.029 | Posttranslational modification |
| Mouse integrase gene (IN) | NF | X52622 | -1.43 | 0.021 | Replication |
| Myogenic differentiation1 (MyoD1) | Mm.1526 | M84918 | -1.52 | 0.032 | Transcription |
| Zinc finger protein 90 (Zfp 90) | Mm.295582 | X79828 | -2.06 | 0.040 | Transcription |
| Histone deacetylase 10 (Hdac 10) | Mm.346413 | AI323 456 | -3.73 | 0.014 | Transcription |
| Prion protein (Prnp) | Mm.648 | M13685 | -1.40 | 0.021 | CNS protein |
| Gene | Mice1 | RT-real time PCR | Microarrays fold difference HBx-LacZ/LacZ | |
| Relative value2 | Fold difference HBx-LacZ/LacZ | |||
| SAA3 | LacZ | 0.37 ± 0.41 | -10.9 | -2.00 |
| HBx-LacZ | 0.03 ± 0.02 | |||
| Creatine Kinase | LacZ | 1.36 ± 1.29 | -1.6 | -1.65 |
| HBx-LacZ | 0.87 ± 0.91 | |||
| MSH6 | LacZ | 1.08 ± 0.64 | 1.5 | 1.50 |
| HBx-LacZ | 1.62 ± 0.64 | |||
| Rb2 | LacZ | 0.47 ± 0.15 | 1.2 | 1.20 |
| HBx-LacZ | 0.59 ± 0.23 | |||
| Peroxiredoxin | LacZ | 0.08 ± 0.02 | 1.5 | 1.40 |
| HBx-LacZ | 0.12 ± 0.08 | |||
As shown in Table 2, the majority of the up-regulated genes in HBx-expressed transgenic mice corresponded to the expression of DNA repair and transmembrane proteins. RAD52 is responsible for DNA double-strand break repair and mitotic recombination and MSH6 for DNA mismatch repair[21]. The transmembrane protein genes implicated in focal adhesion (syndecan 4), viral internalization (CD209), transport of protein between the medial-and trans-Golgi compartment (Golgi SNAP receptor) and in interactions with beta-1 integrin (transmembrane 4 superfamily member 2) were found to be up-regulated. In addition, changes in the expression of the epidermal growth factor and Aven, a caspase inhibitor, were also observed.
Genes whose expression was found to be down-regulated in the liver of HBx-expressed transgenic mice (Table 3) consisted of two main classes: those connected to the transcription control, such as Myogenic differentiation1, Zinc finger protein 90 and Histone deacetylase 10 (HDAC 10), and genes involved in cell metabolism, such as serum amyloid A3 (SAA3), creatine kinase, pepsinogen C, aldolase A, glutathione S-transferase and phospholipase A2. A subclass of down-regulated genes, connected with cholesterol metabolism and isoprenoid synthesis, such as farnesyl diphosphate synthase (FPPS), geranylgeranyl diphosphate synthase (GGPPS) and Cyp7 were also observed. In the group of genes that was found to be down-regulated in the liver of HBx-transgenic mice, we observed several genes like SAA3, aldolase A, creatine kinase, phospholipase A2 that have been reported elsewhere as up-regulated during the course of liver regeneration[22–24].
Interestingly, down-regulation of histone deacetylase and up-regulation of DNA repair genes (Rad52 and MSH6) observed in our study in HBx-transgenic mice after PH have, in agreement with other reports, similar effect on the cell cycle by inducing cell cycle arrest[25–28]. Furthermore, it has previously been reported that HBx expression may interfere with nucleotide excision repair mechanisms[2930] and interact with the DNA repair protein[3132].
In the next subsections we will focus on selected modulated genes implicated in cholesterol regulation and discuss their possible impact on the inhibition of liver regeneration.
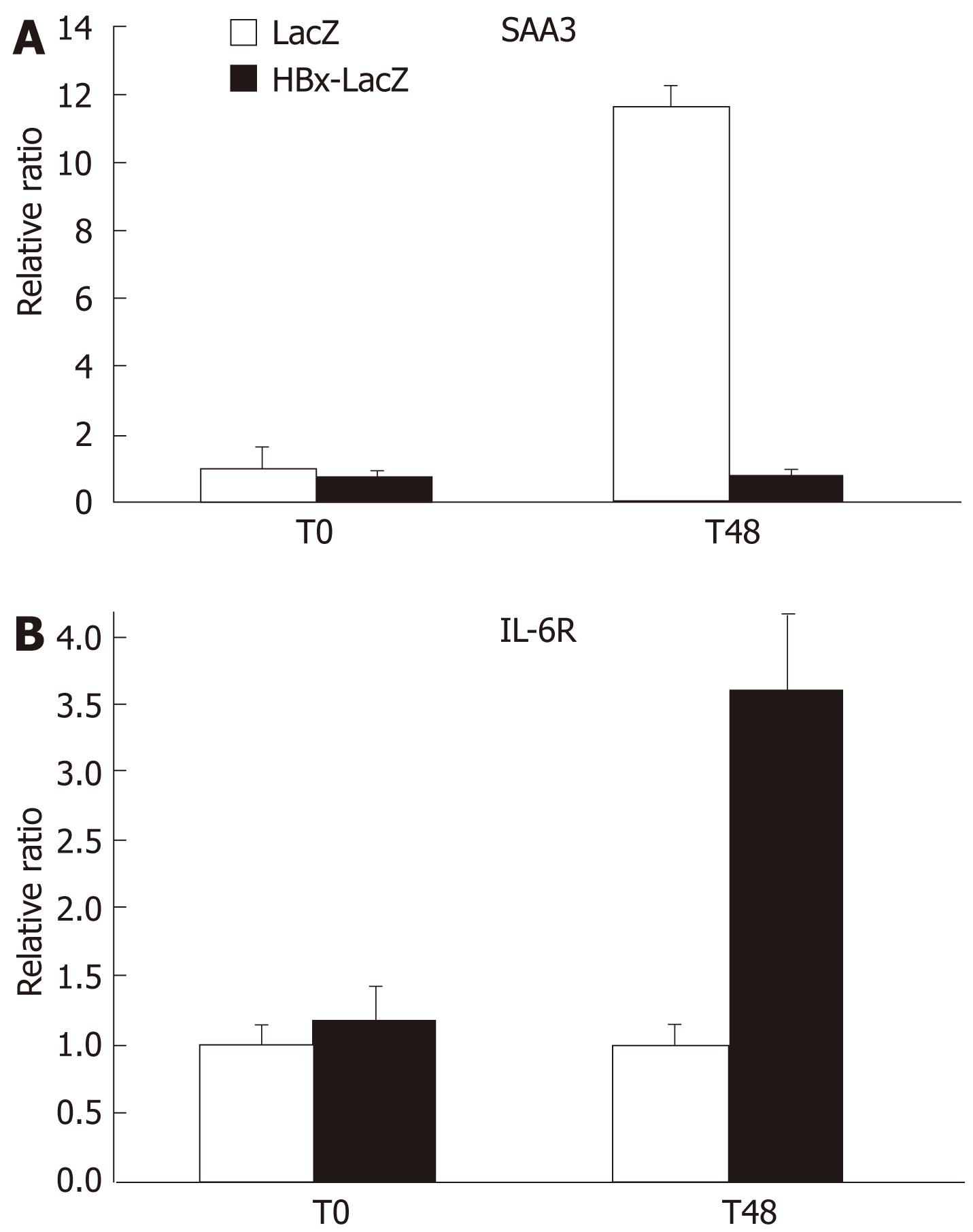
SAA3 is a multifunctional protein involved in acute phase response, inflammation, and cholesterol transport. However, the precise function of SAA in liver regeneration is unclear. The physiological transient induction of SAA appears to be indispensable for the precise regeneration rate after PH[3]. To investigate whether HBx expression was able to completely or partially inhibit SAA3 expression during PH, real time RT-PCR was performed on liver biopsies obtained at the time of PH (T0) and 48 h later from two LacZ or HBx-LacZ transgenic mice. As expected, in the control LacZ animals, a marked increase in SAA expression (more than 10-fold) was observed 48 h after PH as compared to T0 (Figure 1A). By contrast, in HBx-LacZ mice, PH was not associated with an increase in the expression of SAA3 transcript. Thus, HBx induces complete inhibition of SAA response during PH. This may constitute an important factor involved in the impaired liver regeneration previously observed in these animals.
Regulation of SAA expression during the acute phase response depends on hepatic mitogens (Il-6, IL-1, TNF-α)[3]. As the expression of these cytokines is not significantly modified by HBx 48 h after PH, they are probably not implicated in the observed inhibition of SAA3 transcription in HBx-LacZ transgenic mice. To define whether any modulation of the expression of Il-6 receptor (IL-6R), not incorporated in our microarray, is related to the inhibition of SAA expression in HBx-LacZ mice, we used RT-PCR assay to determine the expression of IL-6R in LacZ and HBx-LacZ transgenic mice, at the time of PH and 48 h later. As shown in Figure 1B, prior to PH, no difference in IL-6R expression was observed in HBx-LacZ mice compared to control mice. By contrast, 48 h after PH there was greater than 3-fold up-regulation in HBx-LacZ mice as compared to controls. These findings suggest that HBx is able to modulate Il-6 receptor expression in association with a factor activated during PH and confirms that the lack of SAA response after PH in HBx-expressing transgenic mice is independent of cytokine activity.
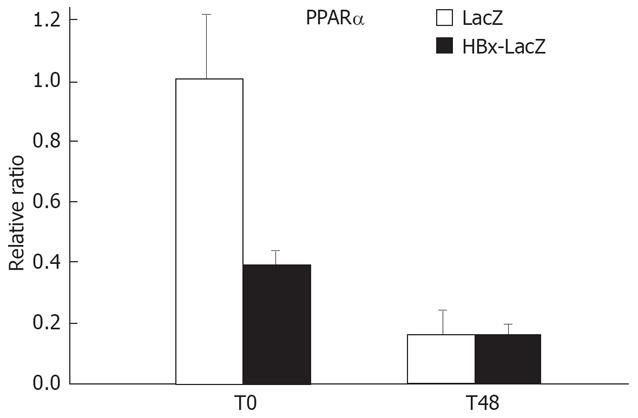
We observed that after PH of HBx-transgenic mice, the expression of enzymes implicated in the isoprenylation biosynthesis (FPPS and GPPS, and to a lesser extent HMG-CoA synthase) was reduced in the liver. Peroxisome proliferator-activated receptor-α (PPARα) is a transcription factor that controls lipid metabolism and glucose homeostasis. PPARα regulates the expression of enzymes implicated in isoprenoid synthesis, and its role in liver regeneration after PH was demonstrated via mechanisms involving prenylation of small GTPases Ras and RhoA independent of TNF-α and IL-6 activation[33–35]. In addition, it was observed that PPARα is involved in the expression of FPPS and HMG-CoA synthase after PH[33]. Thus, we wanted to investigate whether the decrease in the expression of enzymes implicated in isoprenylation observed in HBx-transgenic mice can be explained by a decrease in PPARα expression. We used RT-PCR to assess PPARα expression in LacZ and HBx-LacZ transgenic mice, both at the time of PH and 48 h later. At the time of PH, a 60% decrease in the expression of PPARα gene was observed in HBx-LacZ compared to LacZ transgenic mice (Figure 2). It has been reported that the absence of PPARα expression in mice contributes to impaired liver regeneration[36]. As previously reported, there is a general decrease in PPARα expression after PH[37]. In accordance with the microarray data, there was no difference in PPARα expression between LacZ and HBx-LacZ transgenic mice, 48 h post PH. Thus, the reduced expression of PPARα before PH may be involved in the inhibition of liver regeneration in HBx-LacZ transgenic mice.
In the present study, we used liver biopsies of HBx-expressing transgenic mice, 48 h after partial hepatectomy, to assess the factors implicated in the inhibition of liver regeneration in HBx-transgenic mice, by determining the alterations in mRNA profile. By that means, we extended the results obtained in in vivo studies, demonstrating inhibition of liver regeneration by HBx-expression in HBx-transgenic mice[8]. The present study demonstrates the influence of altered expression of genes involved in transcription control, DNA reparation, cholesterol metabolic pathway and isoprenoid biosynthesis in the reduction of liver mass observed in HBx-transgenic mice[8].
Although the data is controversial, it has been shown that HBx may up-regulate the expression of p21waf1/cip1 and prolong G1→S transition[3839]. A recent study on liver regeneration performed on transgenic mice showed that HBx can block G1/S transition, and so cause the lower liver mass restoration[40]. In this context, we showed that HBx down-regulated HDAC10 and up-regulated DNA repair gene (Rad52 and MSH6) expression, which may have a similar impact on the cell cycle. Indeed, HDAC inhibition causes cell cycle arrest due to an increase in the expression of p21 WAF1/Cip1 and Rb[25264142]. Moreover, it has been reported that Rad52 over-expression affects cell cycle regulation by delaying the exit from G1[27], and that MSH6 protein induces apoptosis by activation of p53[28]. Thus, one of the possible mechanisms that has emerged from our study is in accordance with previously reported activity of HBx, inhibition of cell cycle progression throughout the modulation of pathways implicated in DNA repair and transcription control.
During regeneration induced by tissue loss, a complex of growth factors and cytokines induce hepatocyte progression from G0 to cell cycle to restore the original liver mass. At the same time, Il-6, IL-1 and TNF-α, known to be hepatic mitogens, induce the acute-phase response (APR)[343]. During APR, the hepatic biosynthesis of SAA is up-regulated by proinflammatory cytokines, and the circulating concentration of SAA can increase up to 1000 fold. SAA is induced transiently and returns to its normal low basal level within 72-96 h of the initial stimulus[44]. It has been reported earlier that the mRNA for APR protein such as serum amyloid A increases dramatically during liver regeneration[45]. When we analyzed the changes in the mRNA profile in liver 48 h after PH we also observed a greater than 10-fold increase in the level of SAA mRNA, but only in the control mice. SAA expression level in HBx-transgenic mice 48 h after PH remained unchanged (even slightly lower) when compared to T0. Although the exact function of SAA is still unclear, the physiologic transient induction of SAA appears to be indispensable for proper regeneration after PH. Thus, the lack of SAA response in HBx-transgenic mice after PH may constitute an alternative factor in the impaired regeneration.
The activity of HMG-CoA synthase, farnesyl pyrophosphate synthase and geranylgeranyl pyrophosphate synthase was found to be indispensable for the isoprenylation process and the subsequent cell cycle progression and cell proliferation[3334]. Furthermore, FPPS and GGPPS are implicated in cell cycle progression via their association with the isoprenylation process of GTPases Ras and RhoA proteins[333446]. In addition, PPARα plays an important role in regulating lipid homeostasis. Studies of PPARα null mice have revealed the importance of PPARα in the hepatic lipid metabolism and liver regeneration[3336]. Based on our cDNA microarray results we determined that after PH of HBx-transgenic mice the expression level of these enzymes in the liver is down-regulated. As shown in previous studies, inhibition of enzymes involved in cholesterol and isoprenoid synthesis leads to the upregulation of cdk inhibitors, reduction in cdk, hypophosphorylation of Rb, followed by cell cycle arrest in G1[4748]. Thus, the tempered liver regeneration in HBx-transgenic mice can be explained by the inhibition of the isoprenylation process.
Taken together, our microarray experiments performed on HBx-transgenic mice have provided new mechanisms for HBx-mediated inhibition of liver regeneration. The relevance of our results in the context of HBV-dependent cell cycle inhibition, required for an efficient HBV replication, awaits further evaluation.
The liver has a unique capacity to regenerate after partial hepatectomy (PH) or injury. Experimental evidence supports the hypothesis that proliferation of hepatocytes is responsible for liver regeneration after PH, despite the low replicative rate of hepatocytes in the normal liver. In a previous report we had shown that HBx expression inhibits liver regeneration after PH in HBx expressing transgenic mice. We also demonstrated a paracrine inhibitory effect of HBx on liver cell proliferation. Indeed, transplantation of HBx-expressing liver cells or injection of serum from HBx-transgenic mice into control mice is sufficient to inhibit liver regeneration after PH. Thus, HBx protein may act by combining the intracellular and paracrine effects, and inhibiting the proliferation of both infected and uninfected liver cells in the context of liver regeneration.
Hepatitis B Virus (HBV) infection is a worldwide health problem. It is estimated that 350 million people are chronic carriers of hepatitis B virus (HBV). According to the WHO, hepatitis B is responsible for 1.2 million deaths each year. Chronic HBV infection is a major risk factor for the development of hepatocellular carcinoma. The HBx protein of HBV exhibits pleiotropic effects, that modulate transcription, cell responses to genotoxic stress, protein degradation, cell viability and signalling pathways. Several studies have suggested that HBx protein of HBV, participates in the development of HCC, although the precise role(s) in this process remain unclear.
In the present report, we examined the impact of HBx on the gene expression profile of liver cells in HBx-expressing transgenic mice, 48 h after partial hepatectomy. We used cDNA microarray containing over 5 thousand mouse and rat cDNA elements to determine genes that are altered in the presence of reduced liver regeneration. By demonstrating molecular mechanisms involved in HBx-mediated inhibition of liver regeneration, the data provides a better understanding of the role of the HBx protein in the physiopathology of chronic liver disease caused by HBV infection.
Although these observations were made in the specific context of liver regeneration triggered by partial hepatectomy, they may represent an important mechanism of HBV-dependent liver carcinogenesis in chronic hepatitis and the associated stimulation of liver cell proliferation. Thus, this study may contribute to the development of new approaches based on controlling the cellular effects of viral replication and ultimately provide new tools to control HBV-induced liver disease.
This is a well written, interesting report on gene modulation associated with inhibition of liver regeneration in hepatitis B x protein transgenic mice.
| 1. | Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45-S53. |
| 2. | Albrecht JH, Poon RY, Ahonen CL, Rieland BM, Deng C, Crary GS. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene. 1998;16:2141-2150. |
| 3. | Fulop AK, Pocsik E, Brozik M, Karabelyos C, Kiss A, Novak I, Szalai C, Dobozy O, Falus A. Hepatic regeneration induces transient acute phase reaction: systemic elevation of acute phase reactants and soluble cytokine receptors. Cell Biol Int. 2001;25:585-592. |
| 4. | Arai M, Yokosuka O, Chiba T, Imazeki F, Kato M, Hashida J, Ueda Y, Sugano S, Hashimoto K, Saisho H. Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. J Biol Chem. 2003;278:29813-29818. |
| 5. | Fukuhara Y, Hirasawa A, Li XK, Kawasaki M, Fujino M, Funeshima N, Katsuma S, Shiojima S, Yamada M, Okuyama T. Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol. 2003;38:784-792. |
| 6. | Togo S, Makino H, Kobayashi T, Morita T, Shimizu T, Kubota T, Ichikawa Y, Ishikawa T, Okazaki Y, Hayashizaki Y. Mechanism of liver regeneration after partial hepatectomy using mouse cDNA microarray. J Hepatol. 2004;40:464-471. |
| 7. | Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci USA. 2002;99:11181-11186. |
| 8. | Tralhao JG, Roudier J, Morosan S, Giannini C, Tu H, Goulenok C, Carnot F, Zavala F, Joulin V, Kremsdorf D. Paracrine in vivo inhibitory effects of hepatitis B virus X protein (HBx) on liver cell proliferation: an alternative mechanism of HBx-related pathogenesis. Proc Natl Acad Sci USA. 2002;99:6991-6996. |
| 9. | Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-12734. |
| 10. | Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Brechot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848-4859. |
| 11. | Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, Paterlini-Brechot P, Brechot C, Kremsdorf D. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803-7810. |
| 12. | Anzola M. Hepatocellular carcinoma: role of hepatitis B and hepatitis C viruses proteins in hepatocarcinogenesis. J Viral Hepat. 2004;11:383-393. |
| 13. | Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, Buendia MA. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395-404. |
| 14. | Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Statist. 1996;5:299-314. |
| 15. | Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. |
| 16. | Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet & Mol Bio. 2004;3:Article 3. |
| 17. | Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289-300. |
| 18. | Gervois P, Kleemann R, Pilon A, Percevault F, Koenig W, Staels B, Kooistra T. Global suppression of IL-6-induced acute phase response gene expression after chronic in vivo treatment with the peroxisome proliferator-activated receptor-alpha activator fenofibrate. J Biol Chem. 2004;279:16154-16160. |
| 19. | Kochan Z, Karbowska J. Dehydroepiandrosterone up-regulates resistin gene expression in white adipose tissue. Mol Cell Endocrinol. 2004;218:57-64. |
| 20. | Robert C, McGraw S, Massicotte L, Pravetoni M, Gandolfi F, Sirard MA. Quantification of housekeeping transcript levels during the development of bovine preimplantation embryos. Biol Reprod. 2002;67:1465-1472. |
| 21. | Iaccarino I, Palombo F, Drummond J, Totty NF, Hsuan JJ, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484-486. |
| 22. | Satoh S, Tanaka A, Hatano E, Inomoto T, Iwata S, Kitai T, Shinohara H, Tsunekawa S, Chance B, Yamaoka Y. Energy metabolism and regeneration in transgenic mouse liver expressing creatine kinase after major hepatectomy. Gastroenterology. 1996;110:1166-1174. |
| 23. | Wojtaszek PA, Van Putten V, Nemenoff RA. Activation of a novel form of phospholipase A2 during liver regeneration. FEBS Lett. 1995;367:228-232. |
| 24. | de Jong KP, von Geusau BA, Rottier CA, Bijzet J, Limburg PC, de Vries EG, Fidler V, Slooff MJ. Serum response of hepatocyte growth factor, insulin-like growth factor-I, interleukin-6, and acute phase proteins in patients with colorectal liver metastases treated with partial hepatectomy or cryosurgery. J Hepatol. 2001;34:422-427. |
| 25. | Lavelle D, Chen YH, Hankewych M, DeSimone J. Histone deacetylase inhibitors increase p21(WAF1) and induce apoptosis of human myeloma cell lines independent of decreased IL-6 receptor expression. Am J Hematol. 2001;68:170-178. |
| 26. | Di Padova M, Bruno T, De Nicola F, Iezzi S, D’Angelo C, Gallo R, Nicosia D, Corbi N, Biroccio A, Floridi A. Che-1 arrests human colon carcinoma cell proliferation by displacing HDAC1 from the p21WAF1/CIP1 promoter. J Biol Chem. 2003;278:36496-36504. |
| 27. | Yanez RJ, Porter AC. Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic Acids Res. 2002;30:740-748. |
| 29. | Jia L, Wang XW, Harris CC. Hepatitis B virus X protein inhibits nucleotide excision repair. Int J Cancer. 1999;80:875-879. |
| 30. | Groisman IJ, Koshy R, Henkler F, Groopman JD, Alaoui-Jamali MA. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis. 1999;20:479-483. |
| 31. | Becker SA, Lee TH, Butel JS, Slagle BL. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol. 1998;72:266-272. |
| 32. | Lin-Marq N, Bontron S, Leupin O, Strubin M. Hepatitis B virus X protein interferes with cell viability through interaction with the p127-kDa UV-damaged DNA-binding protein. Virology. 2001;287:266-274. |
| 33. | Wheeler MD, Smutney OM, Check JF, Rusyn I, Schulte-Hermann R, Thurman RG. Impaired Ras membrane association and activation in PPARalpha knockout mice after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2003;284:G302-G312. |
| 34. | Sebti SM. Blocked pathways: FTIs shut down oncogene signals. Oncologist. 2003;8 Suppl 3:30-38. |
| 35. | Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol. 2004;5:355-366. |
| 36. | Anderson SP, Yoon L, Richard EB, Dunn CS, Cattley RC, Corton JC. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology. 2002;36:544-554. |
| 37. | Skrtic S, Carlsson L, Ljungberg A, Linden D, Michalik L, Wahli W, Oscarsson J. Decreased expression of peroxisome proliferator-activated receptor alpha and liver fatty acid binding protein after partial hepatectomy of rats and mice. Liver Int. 2005;25:33-40. |
| 38. | Ahn JY, Jung EY, Kwun HJ, Lee CW, Sung YC, Jang KL. Dual effects of hepatitis B virus X protein on the regulation of cell-cycle control depending on the status of cellular p53. J Gen Virol. 2002;83:2765-2772. |
| 39. | Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 2004;32:2202-2213. |
| 40. | Wu BK, Li CC, Chen HJ, Chang JL, Jeng KS, Chou CK, Hsu MT, Tsai TF. Blocking of G1/S transition and cell death in the regenerating liver of Hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun. 2006;340:916-928. |
| 41. | Siddiqui H, Solomon DA, Gunawardena RW, Wang Y, Knudsen ES. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol Cell Biol. 2003;23:7719-7731. |
| 42. | Wade PA. Transcriptional control at regulatory checkpoints by histone deacetylases: molecular connections between cancer and chromatin. Hum Mol Genet. 2001;10:693-698. |
| 43. | Trautwein C, Rakemann T, Niehof M, Rose-John S, Manns MP. Acute-phase response factor, increased binding, and target gene transcription during liver regeneration. Gastroenterology. 1996;110:1854-1862. |
| 44. | Jensen LE, Whitehead AS. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem J. 1998;334:489-503. |
| 45. | Friedman JM, Chung EY, Darnell JE, Jr . Gene expression during liver regeneration. J Mol Biol. 1984;179:37-53. |
| 46. | Gelb MH. Protein prenylation, et cetera: signal transduction in two dimensions. Science. 1997;275:1750-1751. |
| 47. | Asakage M, Tsuno NH, Kitayama J, Kawai K, Okaji Y, Yazawa K, Kaisaki S, Takahashi K, Nagawa H. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase inhibitor (pravastatin) inhibits endothelial cell proliferation dependent on G1 cell cycle arrest. Anticancer Drugs. 2004;15:625-632. |
| 48. | Fuse M, Tanaka T, Shibata T, Yoshida T, Noguchi Y, Misawa N, Yasuda T, Saito Y, Kohn LD, Tatsuno I. Regulation of geranylgeranyl pyrophosphate synthase in the proliferation of rat FRTL-5 cells: involvement of both cAMP-PKA and PI3-AKT pathways. Biochem Biophys Res Commun. 2004;315:1147-1153. |