Published online Oct 14, 2008. doi: 10.3748/wjg.14.5816
Revised: August 26, 2008
Accepted: September 2, 2008
Published online: October 14, 2008
AIM: To analyze the metastasis-related proteins in hepatocellular carcinoma (HCC) and discover the biomarker candidates for diagnosis and therapeutic intervention of HCC metastasis with bioinformatics tools.
METHODS: Metastasis-related proteins were determined by stable isotope labeling and MS analysis and analyzed with bioinformatics resources, including Phobius, Kyoto encyclopedia of genes and genomes (KEGG), online mendelian inheritance in man (OMIM) and human protein reference database (HPRD).
RESULTS: All the metastasis-related proteins were linked to 83 pathways in KEGG, including MAPK and p53 signal pathways. Protein-protein interaction network showed that all the metastasis-related proteins were categorized into 19 function groups, including cell cycle, apoptosis and signal transduction. OMIM analysis linked these proteins to 186 OMIM entries.
CONCLUSION: Metastasis-related proteins provide HCC cells with biological advantages in cell proliferation, migration and angiogenesis, and facilitate metastasis of HCC cells. The bird’s eye view can reveal a global characteristic of metastasis-related proteins and many differentially expressed proteins can be identified as candidates for diagnosis and treatment of HCC.
- Citation: Song PM, Zhang Y, He YF, Bao HM, Luo JH, Liu YK, Yang PY, Chen X. Bioinformatics analysis of metastasis-related proteins in hepatocellular carcinoma. World J Gastroenterol 2008; 14(38): 5816-5822
- URL: https://www.wjgnet.com/1007-9327/full/v14/i38/5816.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.5816
| Protein name | IPI number | Ratio1 | NO2 | Coverage (%)3 | Category |
| Filamin C | IPI00178352 | 2.49 ± 0.15 | 2 | 1.28 | Cytoskeleton |
| Obscurin | IPI00549822 | 2.91 ± 0.18 | 2 | 1.16 | Cytoskeleton |
| Keratin 8 | IPI00554648 | 0.56 ± 0.15 | 4 | 25.1 | Cytoskeleton |
| Titin | IPI00179357 | 1.73 ± 0.12 | 5 | 1.11 | Cytoskeleton |
| Plectin 7 | IPI00398776 | 2.49 ± 0.13 | 2 | 1.19 | Cytoskeleton |
| MTA2 | IPI00171798 | 0.24 ± 0.11 | 2 | 2.69 | p53 regulation |
| Low density lipoprotein receptor-related protein 1 (LRP1) | IPI00020557 | 0.58 ± 0.15 | 2 | 2.27 | Cell migration, proliferation, angiogenesis |
| Mos | IPI00018290 | 2.86 ± 0.2 | 2 | 6.36 | Mitoses |
| KEGG pathway | IPI accession | Protein description |
| Antigen processing and presentation | IPI00020984 | Calnexin precusor |
| IPI00025252 | Protein disulfide-isomerase A3 precusor | |
| IPI00037070 | Splice isoform 2 of heat shock cognate 71 kDa protein | |
| IPI00144014 | MHC Cass I antigen | |
| IPI00514377 | Heat shock 70 kDa protein | |
| Regulation of actin cytoskeleton | IPI00001814 | Serine/threonine- protein kinase PAK 7 |
| IPI00018290 | Mos | |
| IPI00216691 | Profiline-1 | |
| IPI00384231 | PAK4 protein | |
| Renal cell carcinoma | IPI00001814 | PAK 7 |
| IPI00384231 | PAK4 protein | |
| Thyroid cancer | IPI00022970 | Translocated promoter region |
| Small cell lung cancer | IPI00217461 | Splice isoform 3 of apoptotic protease activating factor 1 |
| IPI00377045 | Laminin alpha 3 splice variant B1 | |
| Prostate cancer | IPI00219757 | Glutahione S-transferase protein |
| p53 pathway | IPI00006160 | Splice isoform alpha of tumor protein P73 |
| IPI00217461 | Splice isoform 3 of apoptotic protease activating factor 1 | |
| MAPK pathway | IPI00018290 | Mos |
| IPI00037070 | Splice isoform 2 of heat shock cognate 71 kDa protein (HSP70) | |
| IPI00155892 | Voltage-dependent calcium channel gamma-5 subunit isoform B | |
| IPI00178352 | Splice isoform 1 of filamin C | |
| IPI00382696 | Splice isoform 1 of filamin B | |
| IPI00514377 | Heat shock 70 kDa protein 1A | |
| ErbB pathway | IPI00001814 | PAK 7 |
| IPI00384231 | PAK4 |
| Cell function | Interactors | Percent (%) |
| Cell cycle | 6 | 0.69 |
| DNA repair | 57 | 6.54 |
| Stress response | 14 | 1.61 |
| Protein biosynthesis | 9 | 1.03 |
| Carbohydrate metabolism | 26 | 2.99 |
| DNA replication | 3 | 0.34 |
| Transcription | 39 | 4.48 |
| RNA processing | 14 | 1.61 |
| RNA localization | 1 | 0.11 |
| Signal transduction | 120 | 13.78 |
| Transport | 41 | 4.71 |
| Protein dephosphorylation | 9 | 1.03 |
| Cell organization and biogenesis | 124 | 14.24 |
| Metabolism | 70 | 8.04 |
| DNA damage response | 3 | 0.34 |
| Protein transport | 59 | 6.77 |
| Protein phosphorylation | 60 | 6.89 |
| Protein degradation | 37 | 4.25 |
| DNA repair | 13 | 1.49 |
| Unknown | 166 | 19.06 |
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world with 626 000 new cases occurred in 2002[1]. Surgical resection remains the primary treatment of choice, but because of the very high rate of metastasis and poor prognosis, the number of deaths is almost the same of new cases (598 000) occurred each year worldwide. Metastasis is a highly complicated biological process involving multiple proteins functioning in a coordinated manner and the molecular mechanism underlying the metastasis of HCC is not completely understood. Investigations on metastasis-related proteins and the molecular mechanism are urgent in the campaign against HCC both in the world and in China[2]. Technology advances in genomics and proteomics would facilitate our understanding of HCC metastasis. Genome analysis with DNA array can be used to scan different gene expressions in different HCC samples and valuable biomarkers have been discovered[3]. However, no well linear correlation has been found between gene and protein expression levels due to splice process of mRNA and post-transcriptional regulation[4,5]. Hence, differential expression profile analysis of a large number of proteins is an essential step in understanding the mechanism of metastasis and in discovering the diagnostic markers and therapeutic targets for HCC.
Quantitative proteomics using stable isotope labeling has the necessary ability to rapidly identify and quantify differentially expressed proteins in two or more samples with a high throughout, and thus is currently used as the workhorse for discovering and validating proteins related with a special disease in clinical research. In our previous work, the proteome profiles of two HCC cell lines with different metastasis potentials were compared using stable isotope labeling and 223 differentially expressed proteins were identified confidently. In this study, we analyzed these proteins with bioinformatics tools to discover their biological role in the process of metastasis. Our results show that bioinformatics analysis can provide a valuable molecular basis for systematic interpretation of the mechanism underlying HCC metastasis where potential protein markers could be characterized.
[5,5,5-d3] leucine (leu-d3) was purchased from Cambridge Isotope (Andover, MA). Formic acid (FA) and trifluoroacetic acid (TFA) were from FlukA (Switzerland) and acetonitrile (ACN) was from Merk (Darmastadt, Germany). Trypsins were purchased form Promega (Madison, WI). Dulbecco’s modified Eagle’s medium (DMEM) and dialyzed fetal bovine serum (FBS) were from Gibco-Invitrogen (Grand Island, NY, USA), and normal FBS was from PAA Laboratories GmbH. All others components of cell culture medium, amino acid kits, inorganic salts, vitamin solution, etc, were obtained from Sigma (St, Louis, MO). Other chemicals for SDS-PAGE gel electrophoresis, peptide extraction, and sample preparation for LC-ESI-MS were purchased from Sigma (St. Louis, MO, USA).
HCC cell line MHCC97H with a high metastasis potential was cultured in a normal DMEM, while MHCC97L with a low metastasis potential was cultured in a special DMEM, in which leu-d3 was supplemented to substitute its unlabeled counterpart depleted in the normal DMEM[6]. Cells were harvested at the 80% confluence and treated with a lysis buffer containing 20 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 3 mmol/L MgCl2, 1 mmol/L CaCl2, 10 mL/L NP-40, and 1 mmol/L PMSF. Samples were centrifuged at 10 000 r/min for 30 min at 4°C and stored at -80°C for electrophoresis. Samples from MHCC97H and MHCC97L were mixed at 1:1 of total protein mass and separated on SDS-PAGE. Trypsin digestion and peptide extraction were carried out as previously described[6].
Peptides were resuspended in 5 μL of 1 mL/L TFA solution and directly loaded onto a 300 μm × 100 mm C18 column (Grace Vydac, Hesperia, CA) using an Agilent 1100 binary pump. The following gradients were used for peptide separation: 50-400 ml/L B from 0 min to 50 min, 400-800 ml/L B from 50 min to 70 min, 800-950 ml/L B from 70 min to 90 min, where solvent A was 1 mL/L FA and solvent B was 1 mL/L FA in ACN. Fractions were collected and deposited on a 12 × 16 array Applied Biosystems plate. Alpha-cyano-4-hydroxycinnamic acid (CHCA) was used as a matrix. Both MALDI-MS and MS/MS mass spectra were obtained with the Applied Biosystems 4700 proteomics analyzer (Framingham, MA, USA). MS/MS data were searched from the international protein index (IPI, version 3.07) human database with Mascot (version 1.9, Matrix Science, Boston, MA). Leu-d3 modification was added into a modification file. Error tolerance of peptide MS was set at 0.3 U, MS/MS tolerance at 0.6 U. One missed cleavage per peptide was allowed.
UniProt (Swiss-Prot) format file (ipi.HUMAN.dat) of IPI entries and cross-reference file were downloaded from IPI database (human, version 3.07, ftp://ftp.ebi.ac.uk/pub/databases/IPI/). GeneID, gene symbol and Swiss-Prot accession number for each metastasis-related protein were retried from the cross-reference file. Amino acid sequences of IPI entries in ipi.HUMAN.dat were used in prediction of transmembrane segments with Phobius (http://www.ebi.ac.uk/Tools/phobius/)[7].
Protein-protein interactions were extracted from human protein reference Database (HPRD)[8] and interaction network was demonstrated with Osprey (version 1.2.0, http://biodata.mshri.on.ca/osprey/index.html)[9]. GeneIDs were mapped to the KEGG pathway with a Perl script through the API service at the KEGG (http://www.genome.jp/kegg/soap/doc/keggapi_manual.html)[10].
Cross-references to each entry in the online mendelian inheritance in man (OMIM) database were obtained from the UniProtKB/Swiss-Prot subset of IPI entries through a Perl script. The detailed descriptions of OMIM entries were obtained from the OMIM web site (http://www.ncbi.nlm.nih.gov/omim).
Perl scripts were written in the environment of ActivePerl (version 5.8.8, http://www.activeperl.com).
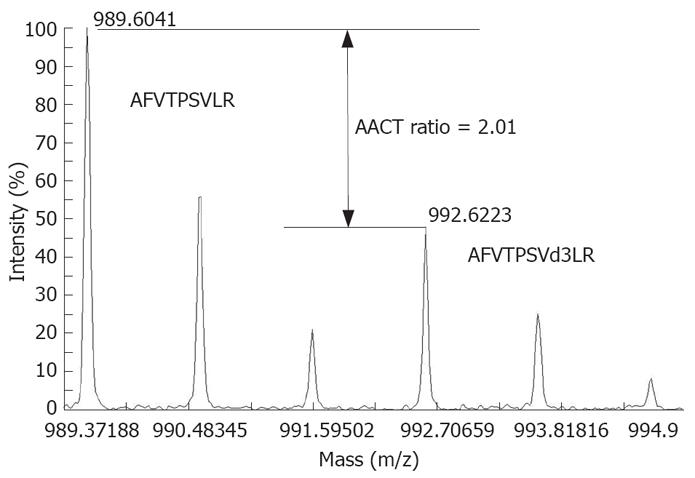
HCC cell lines MHCC97H and MHCC97L were subcloned from the same cell line MHCC97, with a high and low metastasis potential, respectively[11]. MHCC97L was cultured in a special medium for 10 passages and thus labeled completely with leu-d3 while MHCC97H was cultured in a normal medium. Samples from normal (MHCC97H) and leu-d3-labeled (MHCC97L) cells were combined and analyzed with off-line LC-MALDI-MS. Changes in expression of 506 proteins were quantified as previously described[12]. A high confidence as a single or multiple leucine-containing tryptic peptides was detected for each of these proteins. Peptide AFVTPSVLR from Pr-domain zinc finger protein 5 (IPI00032997) contained one leucine, and the expression ratio was quantified as 2.01:1 (MHCC97H: MHCC97L) by comparing the intensity of peaks 989.6 and 992.6 (Figure 1). The results will be reported in another article in details and some important proteins are listed in Table 1. A total of 223 proteins with an expression ratio > 1.5 or < 0.67 were defined as metastasis-related proteins and further analyzed with bioinformatics tools.
The use of 2DE-MS for profiling the membrane proteins is limited due to the relative insolubility of these proteins under conditions suitable for 2-DE analysis[13,14]. However, membrane proteins are likely to be very import in understanding metastasis. For this reason, we undertook SDS-PAGE to identify more membrane proteins in our study. Phobius is a web tool to predict transmembrane topology of protein with a hidden Markov model[7]. Therefore, it was used to predict the transmembrane segments of metastasis-related proteins in our study. Several membrane proteins with multi-transmembrane segments were identified, such as sodium channel protein type 1 subunit alpha (IPI00216029) containing 24 transmembrane segments, brain calcium channel I (IPI00217499) containing 23 segments, HH1 (IPI00377006) containing 22 segments, hNaN (IPI00513973) containing 20 segments. It was difficult to detect these proteins with multi-transmembrane segments using the 2DE method due to their low solubility in 2DE lysis buffer. In our study, more membrane proteins were detected than in a previous report using 2DE[15]. The dataset of metastasis-related proteins here provided comprehensive biology information on the molecular mechanism of HCC and would facilitate the discovery of biomarkers for its diagnosis and therapy.
Based on the API service of KEGG (http://www.genome.jp/kegg/soap/doc/keggapi_manual.html)[10], a Perl script was used to associate these metastasis-related proteins with known pathways and disease states (Table 2). All the proteins were linked to 83 biological pathways and human diseases in the KEGG, including antigen processing and presentation, regulation of actin cytoskeleton and signal pathways. Among these pathways, many biological processes were connected with metastasis in previous studies, including MAPK, p53 and ErbB, which are involved in regulating cell proliferation, angiogenesis and migration. In addition, several metastasis-related proteins were suggested to be involved in other types of cancer. For example, PAK7 and PAK 4 functioned in regulation of renal cell carcinoma according to the annotations in the KEGG.
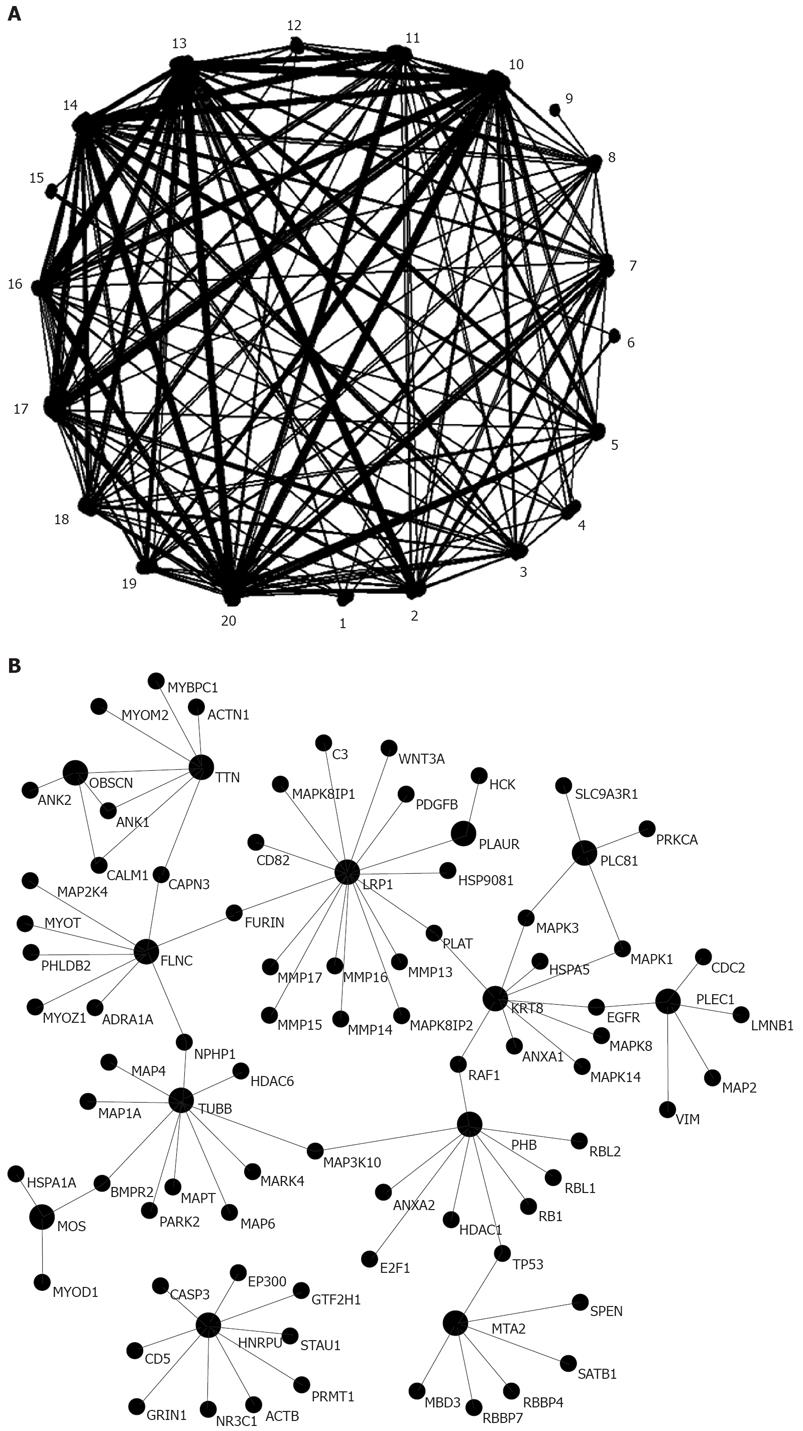
With the software Osprey and the protein-protein interaction data from HPRD, protein-protein interaction network of metastasis-related proteins was visualized (Figure 2, Table 3). The network contained 871 proteins (nodes) and 892 interactions (edges), and the edge/node was 1.02. Osprey also provides a functional tool to cluster proteins by their GO process, and proteins sharing the same GO process were grouped together. Based on the GO annotations in Osprey, all the proteins in the network were classed into 19 known biology function groups. For example, 120 proteins were involved in signal transduction, 57 in DNA repair, and 6 in cell cycle (Table 3). This network could facilitate us to pinpoint the key proteins that play an important role in HCC metastasis and also help us to identify potential markers for the diagnosis and treatment of HCC.
OMIM is one of the most comprehensive resources of human genes and genetic disorders to support research in human genomics and proteomics[16]. A total of 145 proteins were linked to 186 OMIM entries, of which 141 described disease genes and 45 disease phenotypes. Among the disease phenotypes, one was cirrhosis (OMIM #215600) which would afford the development of HCC.
The accuracy of comparative proteomics using stable isotope labeling can be affected by various factors, such as isotope distribution[17]. When the peptide contains one leucine, M+ ion peak of labeled peptide overlaps with the isotope distribution of the corresponding normal peptide. In order to eliminate the effect of isotope distribution and ensure the accuracy of quantification, the theoretical intensity of (M+3)+ peak of normal peptide is subtracted from the labeled peak by calculating its isotope distribution[12]. Peaks with a signal-to-noise ratio < 30 are also excluded in quantification to attenuate the effect of noise on quantification. These modifications ensure the reliability of quantification with stable isotope labeling.
It was reported that some differentially expressed proteins function in the process of metastasis and plectin has an essential role in cell migration through MAP kinase signaling cascades[18,19]. Filamin C can organize actin polymerization and its dysregulation has been observed in some cancer samples[20]. Obscurin binding to titin and sarcomeric myosin[21,22] plays a role in the regular alignment of network sarcoplasmic reticulum developing sarcomeres[23]. The gene encoding this protein is interrupted by the translocation in Wilms’ tumor and functions as a tumor suppressor[24]. Another study showed that obscurin is involved in gastrointestinal stromal tumor and leiomyosarcomas[25]. Proto-oncogene serine/threonine-protein kinase Mos acts as an upstream activator of the MEK/MAPK/p90Rsk pathway regulating M-phase and G2 arrest[26,27]. LRP1 can mediate growth inhibition by IGFBP-3 and cell migration inhibition by binding to apoE[28]. LRP1-mediated clearance of uPA is one of the mechanisms involved in the control of human thyroid carcinoma cell invasion[29]. It was reported that up-regulation of matrix metalloproteinase-9 by tPA in cell culture and in vivo is mediated by LRP1[30]. LRP1 also plays a role in determining the blood vessel structure and angiogenesis[31].
Two biological pathways discovered by KEGG analysis in our study were dysregulated and connected with HCC metastasis. The tumor suppressor protein p53 plays a pivotal role in the regulation of apoptosis and cell cycle arrest. In our study, two p53 regulators, p73 and apoptotic protease activating factor 1, were differentially expressed. MTA2 is a p53-interacting protein that induces p53 deacetylation[32,33]. Dysregulation of p53 function is linked with an unfavorable prognosis of a large number of more aggressive tumor types[34]. Eukaryotic cells possessing multiple MAPK pathways coordinately regulate diverse cellular activities including motility, survival, apoptosis and differentiation[35,36]. The Mos/MAPK/p90Rsk pathway regulates cell cycle progression in oocytes[37], whereas ectopic Mos expression in the early cleavage embryo induces M phase arrest[27]. HSP70 exhibits regulatory functions of c-Jun, ERK and the JNK pathway, thus inhibiting cell apoptosis[38]. The MAPK signaling pathway has long been identified as a convergence point for normal and pathologic signaling inputs, rendering it an appealing target for therapeutic intervention[39]. Various treatment modalities targeting p53 and MAPK pathway are currently under investigation, and dysregulation of p53 and MAPK pathway in HCC metastasis would facilitate finding targets for HCC therapy[39-41].
In conclusion, metastasis-related proteins are dysregulated in HCC metastasis. Biochemical alterations in cell proliferation and migration, angiogenesis and immune response confer selective biological advantages to HCC cells in the process of metastasis. Bioinformatics analysis of metastasis- related proteins provides valuable biological information on the molecular mechanism of metastasis and potential therapeutic targets for HCC.
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world with a high death rate. Metastasis is the major cause of HCC-related death. Discovering metastasis-related proteins would facilitate the diagnosis and treatment of HCC. Quantitative proteomics with stable isotope labeling is a powerful tool to analyze proteome differences between samples with different metastasis and to discover potential therapeutic targets for HCC.
Our study showed the differential proteome profiles of two HCC cell lines with metastasis using stable isotope labeling. Based on the functional annotations with bioinformatics tools, metastasis-related proteins were functionally annotated with Kyoto encyclopedia of genes and genomes (KEGG) pathway, protein-protein interactions from human protein reference database (HPRD) and diseases from online mendelian inheritance in man (OMIM). Functional annotations showed that many proteins in the profile were clearly connected with the process of tumor metastasis.
To the best of our knowledge, the present study showed the largest differential proteome profile of HCC metastasis. Functional annotations with bioinformatics tools showed that metastasis-related proteins were linked with 82 KEGG pathways, 892 interactions and 186 disease entries in OMIM, suggesting that they play a possible role in metastasis of HCC.
The differential proteome profile gives more valuable information on the molecular mechanism of metastasis of HCC and provides potential biomarkers for the diagnosis and treatment ofr HCC.
This is a well conducted study. The manuscript describes the differential proteome profile that gives more information on the molecular mechanism of metastasis of HCC. The study also invested certain potential biomarkers that can be used in the diagnosis and treatment of HCC.
Peer reviewer: Dr. Xin-Yuan Guan, Department of Clinical Oncology, University of Hong Kong, Room 109, Estate Building, 10 Sassoon Road, Hong Kong 852, China
S- Editor Zhong XY L- Editor Wang XL E- Editor Yin DH
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. |
| 2. | Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH, Wang L, Zhou J, Qiu SJ, Li Y. A decade's studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:187-196. |
| 3. | Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J, Xue Q. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol. 2003;129:43-51. |
| 4. | Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720-1730. |
| 5. | Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357-7368. |
| 6. | Shui W, Liu Y, Fan H, Bao H, Liang S, Yang P, Chen X. Enhancing TOF/TOF-based de novo sequencing capability for high throughput protein identification with amino acid-coded mass tagging. J Proteome Res. 2005;4:83-90. |
| 7. | Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429-W432. |
| 8. | Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363-2371. |
| 9. | Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. |
| 10. | Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480-D484. |
| 11. | Li Y, Tang ZY, Ye SL, Liu YK, Chen J, Xue Q, Chen J, Gao DM, Bao WH. Establishment of cell clones with different metastatic potential from the metastatic hepatocellular carcinoma cell line MHCC97. World J Gastroenterol. 2001;7:630-636. |
| 12. | Gu S, Liu Z, Pan S, Jiang Z, Lu H, Amit O, Bradbury EM, Hu CA, Chen X. Global investigation of p53-induced apoptosis through quantitative proteomic profiling using comparative amino acid-coded tagging. Mol Cell Proteomics. 2004;3:998-1008. |
| 13. | Carter P, Smith L, Ryan M. Identification and validation of cell surface antigens for antibody targeting in oncology. Endocr Relat Cancer. 2004;11:659-687. |
| 14. | Flory MR, Griffin TJ, Martin D, Aebersold R. Advances in quantitative proteomics using stable isotope tags. Trends Biotechnol. 2002;20:S23-S29. |
| 15. | Ding SJ, Li Y, Shao XX, Zhou H, Zeng R, Tang ZY, Xia QC. Proteome analysis of hepatocellular carcinoma cell strains, MHCC97-H and MHCC97-L, with different metastasis potentials. Proteomics. 2004;4:982-994. |
| 16. | Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514-D517. |
| 17. | Thiede B, Kretschmer A, Rudel T. Quantitative proteome analysis of CD95 (Fas/Apo-1)-induced apoptosis by stable isotope labeling with amino acids in cell culture, 2-DE and MALDI-MS. Proteomics. 2006;6:614-622. |
| 18. | Osmanagic-Myers S, Gregor M, Walko G, Burgstaller G, Reipert S, Wiche G. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J Cell Biol. 2006;174:557-568. |
| 19. | Boczonadi V, McInroy L, Maatta A. Cytolinker cross-talk: periplakin N-terminus interacts with plectin to regulate keratin organisation and epithelial migration. Exp Cell Res. 2007;313:3579-3591. |
| 20. | Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645-6650. |
| 21. | Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065-1072. |
| 22. | Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am J Physiol Cell Physiol. 2004;287:C209-C217. |
| 23. | Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, Russell MW, Bloch RJ. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006;20:2102-2111. |
| 24. | Vernon EG, Malik K, Reynolds P, Powlesland R, Dallosso AR, Jackson S, Henthorn K, Green ED, Brown KW. The parathyroid hormone-responsive B1 gene is interrupted by a t(1;7)(q42;p15) breakpoint associated with Wilms' tumour. Oncogene. 2003;22:1371-1380. |
| 25. | Price ND, Trent J, El-Naggar AK, Cogdell D, Taylor E, Hunt KK, Pollock RE, Hood L, Shmulevich I, Zhang W. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad Sci USA. 2007;104:3414-3419. |
| 26. | Peter M, Labbe JC, Doree M, Mandart E. A new role for Mos in Xenopus oocyte maturation: targeting Myt1 independently of MAPK. Development. 2002;129:2129-2139. |
| 27. | Wu JQ, Hansen DV, Guo Y, Wang MZ, Tang W, Freel CD, Tung JJ, Jackson PK, Kornbluth S. Control of Emi2 activity and stability through Mos-mediated recruitment of PP2A. Proc Natl Acad Sci USA. 2007;104:16564-16569. |
| 28. | Zhu Y, Hui DY. Apolipoprotein E binding to low density lipoprotein receptor-related protein-1 inhibits cell migration via activation of cAMP-dependent protein kinase A. J Biol Chem. 2003;278:36257-36263. |
| 29. | Sid B, Dedieu S, Delorme N, Sartelet H, Rath GM, Bellon G, Martiny L. Human thyroid carcinoma cell invasion is controlled by the low density lipoprotein receptor-related protein-mediated clearance of urokinase plasminogen activator. Int J Biochem Cell Biol. 2006;38:1729-1740. |
| 30. | Wang X, Lee SR, Arai K, Lee SR, Tsuji K, Rebeck GW, Lo EH. Lipoprotein receptor-mediated induction of matrix metalloproteinase by tissue plasminogen activator. Nat Med. 2003;9:1313-1317. |
| 31. | Gaultier A, Salicioni AM, Arandjelovic S, Gonias SL. Regulation of the composition of the extracellular matrix by low density lipoprotein receptor-related protein-1: activities based on regulation of mRNA expression. J Biol Chem. 2006;281:7332-7340. |
| 32. | Yao YL, Yang WM. The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J Biol Chem. 2003;278:42560-42568. |
| 33. | Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377-381. |
| 34. | Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027-1036. |
| 35. | Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505-516. |
| 36. | Huang D, Ding Y, Luo WM, Bender S, Qian CN, Kort E, Zhang ZF, VandenBeldt K, Duesbery NS, Resau JH. Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Res. 2008;68:81-88. |
| 37. | Inoue D, Ohe M, Kanemori Y, Nobui T, Sagata N. A direct link of the Mos-MAPK pathway to Erp1/Emi2 in meiotic arrest of Xenopus laevis eggs. Nature. 2007;446:1100-1104. |
| 38. | Lee JS, Lee JJ, Seo JS. HSP70 deficiency results in activation of c-Jun N-terminal Kinase, extracellular signal-regulated kinase, and caspase-3 in hyperosmolarity-induced apoptosis. J Biol Chem. 2005;280:6634-6641. |
| 39. | Papatsoris AG, Karamouzis MV, Papavassiliou AG. The power and promise of "rewiring" the mitogen-activated protein kinase network in prostate cancer therapeutics. Mol Cancer Ther. 2007;6:811-819. |
| 40. | Bell HS, Ryan KM. iASPP inhibition: increased options in targeting the p53 family for cancer therapy. Cancer Res. 2008;68:4959-4962. |