Published online Jan 21, 2008. doi: 10.3748/wjg.14.479
Revised: August 30, 2007
Published online: January 21, 2008
AIM: To compare the clinical performance of a real-time PCR assay with the COBAS Amplicor Hepatitis B Virus (HBV) Monitor test for quantitation of HBV DNA in serum samples.
METHODS: The reference sera of the Chinese National Institute for the Control of Pharmaceutical and Biological Products and the National Center for Clinical Laboratories of China, and 158 clinical serum samples were used in this study. The linearity, accuracy, reproducibility, assay time, and costs of the real-time PCR were evaluated and compared with those of the Cobas Amplicor test.
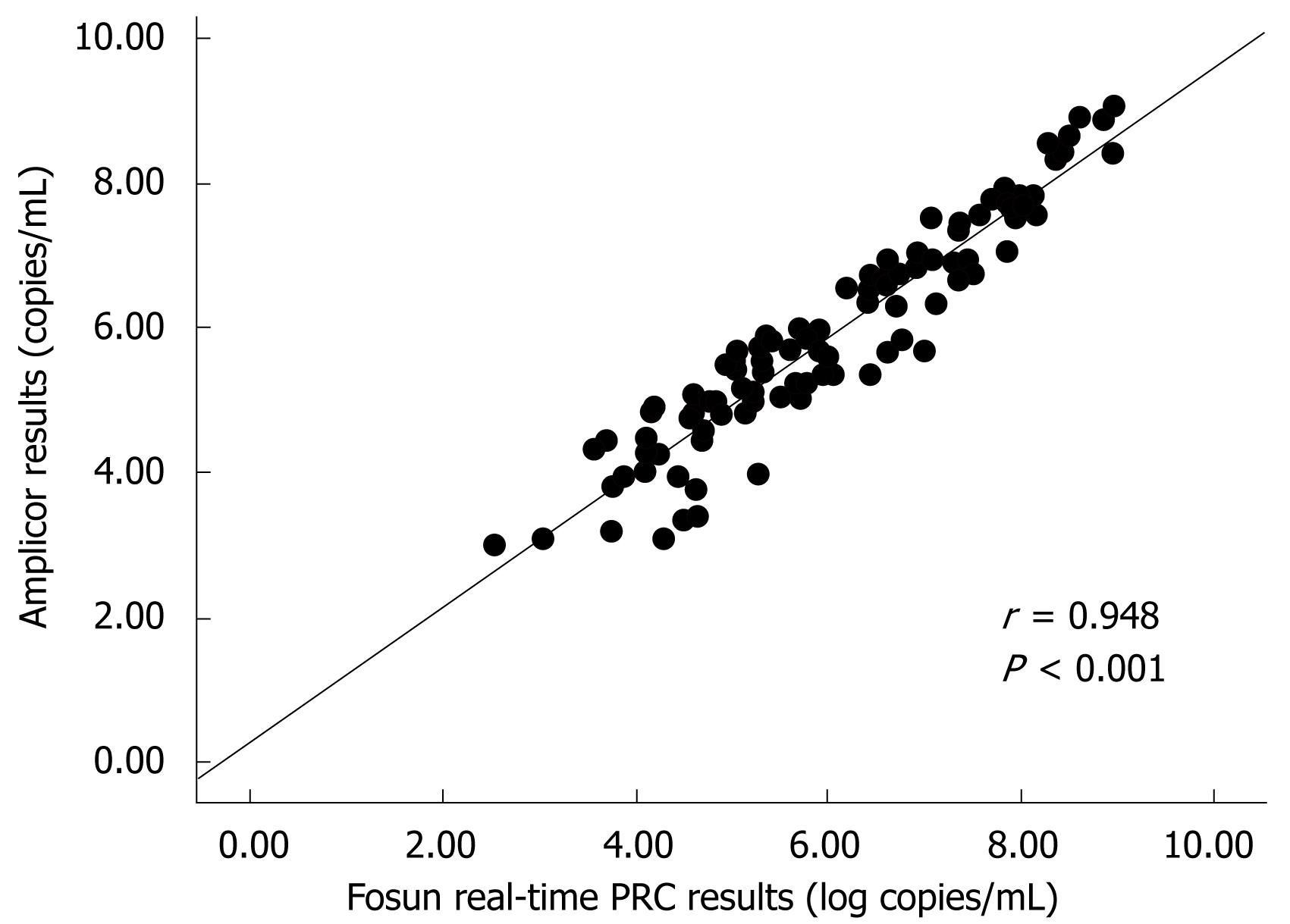
RESULTS: The intra-assay and inter-assay variations of the real-time PCR ranged from 0.3% to 3.8% and 1.4% to 8.1%, respectively. The HBV DNA levels measured by the real-time PCR correlated very well with those obtained with the COBAS Amplicor test (r = 0.948). The real-time PCR HBV DNA kit was much cheaper and had a wider dynamic range.
CONCLUSION: The real-time PCR assay is an excellent tool for monitoring of HBV DNA levels in patients with chronic hepatitis B.
- Citation: Shi M, Zhang Y, Zhu YH, Zhang J, Xu WJ. Comparison of real-time polymerase chain reaction with the COBAS Amplicor test for quantitation of hepatitis B virus DNA in serum samples. World J Gastroenterol 2008; 14(3): 479-483
- URL: https://www.wjgnet.com/1007-9327/full/v14/i3/479.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.479
It is estimated that 350 million individuals are chronically infected with hepatitis B virus (HBV) worldwide, and about one-third of these live in China[1]. HBV infection can cause chronic hepatitis, cirrhosis and hepatocellular carcinoma[2]. Among patients with active viral replication, cirrhosis will develop in 15%-20% within 5 year[3], and 70-90 percent of cases of hepatocellular carcinoma occur against a background of cirrhosis[4]. Quantitation of HBV DNA in serum or plasma has been an important tool in identifying individuals with active hepatitis B, to monitor the efficiency of antiviral treatment, and to predict whether the treatment will be successful[5–9].
Several commercial molecular assays have been developed for quantitation of HBV DNA in serum or plasma samples. One of these assays is the COBAS Amplicor HBV Monitor, which is based on the amplification of DNA targets by the use of HBV-specific primers. Other methods, such as the VERSANT HBV DNA 3.0 assay, which is based on branched-DNA signal amplification, and Hybrid Capture II, which is based on hybridization of a chemiluminescent probe, are also routinely used in diagnostic laboratories[1011]. However, the costs of these assays are too high to be used in developing countries such as China, and in these countries the HBV burden is much heavier than in developed countries.
Recently, quantitative real-time PCR has been used to detect and quantify HBV levels in serum or plasma samples. This assay is based on the relationship between the initial DNA target levels and the threshold cycle (Ct) of the amplification products. The Ct value is smaller when the initial DNA target level is higher. Several evaluation studies have shown that real-time PCR has a higher sensitivity, a broader dynamic range, and an accurate quantitation of HBV DNA compared to those of the existing commercial methods[12–15].
The Fosun real-time PCR HBV kit is a commercial assay for quantitation of serum HBV DNA levels based on TaqMan PCR technology. It has been approved by the State Food and Drug Administration (SFDA) of China for in vitro diagnostic use. This assay is widely used in Chinese laboratories for quantitative detection of serum HBV levels in patients with HBV infection. However, the clinical performance of this real-time PCR assay has not been compared with that of commercial assays routinely used in the United States and Europe. The aim of this study was to evaluate the clinical performance of the Fosun real-time PCR HBV assay on the ABI 7500 sequence detector for the quantitative detection of HBV DNA in serum samples. The results were compared to those obtained with the same samples using the COBAS Amplicor HBV Monitor system.
Serum samples from 158 individuals, including 118 patients with chronic hepatitis B (46 genotype B, 65 genotype C, and 7 mixed genotype B and C), and 40 blood donors, were analyzed in parallel by the Fosun real-time PCR HBV kit (Fosun Diagnostics, Shanghai, China) and COBAS Amplicor HBV Monitor (Roche Diagnostics). All the patients were positive for hepatitis B surface antigen (HBsAg) for at least 6 mo, and were negative for antibodies to hepatitis A, C, D and E viruses. None of these patients had received any antiviral treatment at the time of serum collection.
An HBV reference serum from the National Center for Clinical Laboratories of China with a DNA level of 7.5 × 108 copies/mL was used to determine the linearity and detection limit of the real-time PCR. This reference serum was 10-fold serially diluted into a spectrum of reference samples with HBV DNA levels ranging from 7.5 × 101 to 7.5 × 108 copies/mL.
A set of HBV reference sera established by the Chinese National Institute for the Control of Pharmaceutical and Biological Products (NICPBP), with HBV DNA concentrations ranging from < 1000 to 6.17 × 108 copies/mL of genotype C were used for standardization.
For Fosun real-time PCR, HBV DNA was extracted from 100 &mgr;L serum by using the extraction reagents included in the kit, in accordance with the manufacturer’s instructions. The amplification was performed on an ABI 7500 sequence detector (Applied Biosystems, Foster City, CA, USA) by incubating the reaction mixture (50 &mgr;L) at 50°C for 2 min, 94°C for 5 min, followed by 40 cycles of PCR amplification at 93°C for 30 s and 60°C for 90 s. The Ct value was defined as the number of fractional cycles in which the fluorescence emitted by the reaction mixture exceeded a preset threshold, and marked the beginning of an exponential growth of the fluorescence signal. The results were analyzed with ABI Prism software (Applied Biosystems). A four-point external standard set was used to calculate the initial copy number of the samples. The dynamic range of Fosun real-time PCR HBV DNA kits, as stated by the manufacturer, is 4.2 × 102- 7.5 × 108 copies/mL.
Amplicor tests were performed by using the COBAS Amplicor system and the AMPLICOR HBV Monitor assay according to manufacturer’s instructions. The highly conserved HBV precore/core region and the internal standard DNA were amplified in the same well, but hybridized in separate wells. The amount of HBV DNA was calculated from the ratio of the HBV well to the internal standard well, and the copy number per milliliter was calculated from a standard curve. The dynamic range of the Amplicor test was 3 × 102-2 × 105 copies/mL.
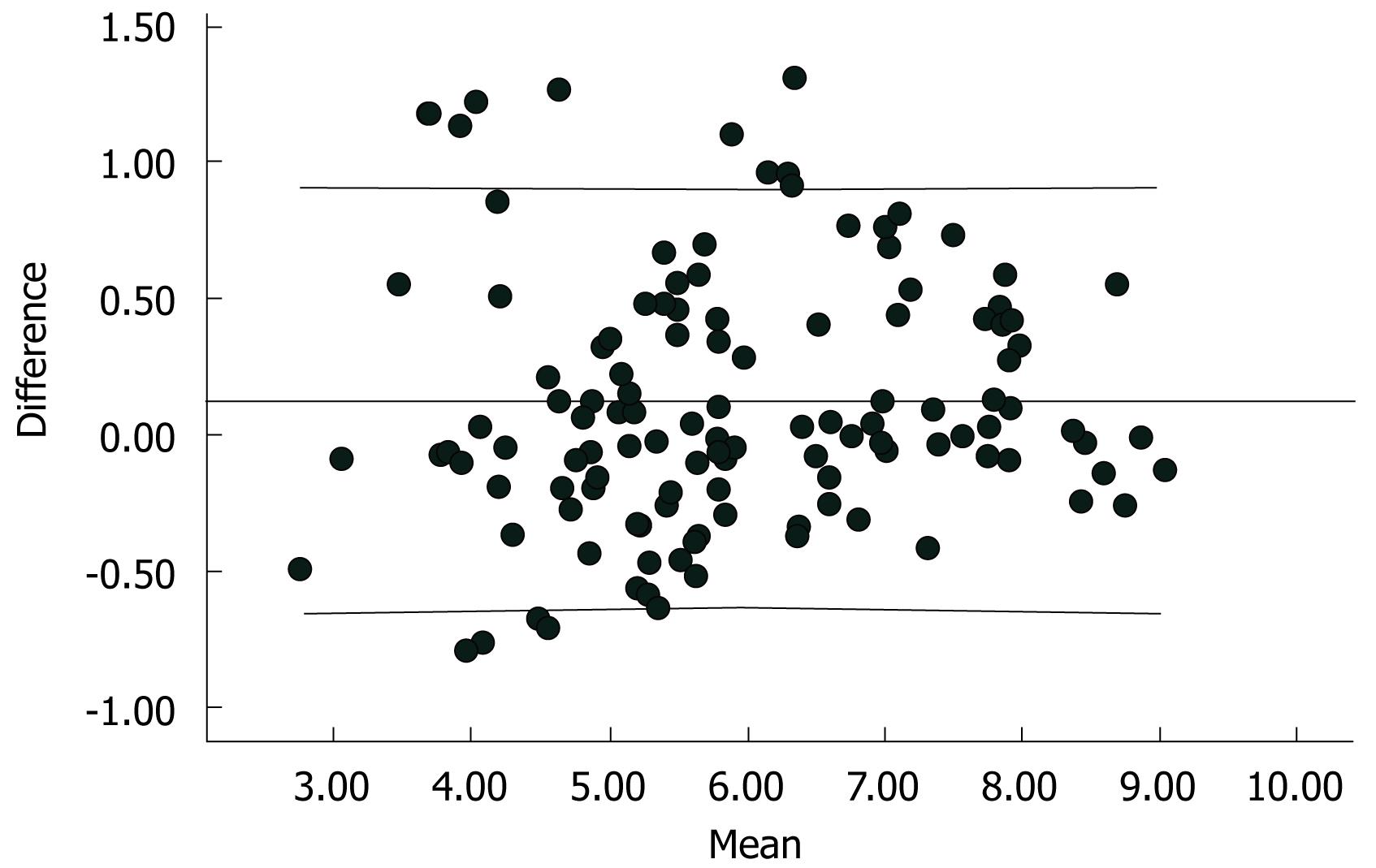
Statistical analyses were performed using the Statistical Program for Social Sciences (SPSS 13.0 for Windows; SPSS, Chicago, IL, USA). Correlation between the quantitative results from the two assays was determined with scatter plots and Spearman’s coefficient analysis, after logarithmic transformation of data with skewed distribution. The Bland-Altman method was used to assess the agreement between the values obtained with the two assays[16].
The real-time PCR was able to detect seven (87.5%) of the eight dilutions of the reference serum. The tested values and the normal values of the dilutions were well matched (r = 0.999, P < 0.001). The converting formula was:
Log (HBV DNA level by real-time PCR) = 1.098 [Log (Reference HBV DNA level)] - 0.735
The real-time PCR was able to detect HBV DNA levels as low as 750 copies/mL, which is similar to the detection limit claimed by the manufacturer.
To determine the dynamics and accuracy of the Fosun real-time PCR kit, the reference serum of the Chinese NICPBP was tested three times with kits with different lot numbers. Fosun real-time PCR kits detected all seven dilutions of the reference serum, and all the quantitative results were within the reference ranges (Table 1). The inter-assay variation ranged from 0.3% to 3.8%. The detection limit of the real-time PCR was 442 copies/mL.
| NICPBP references | Reference range (copies/mL) | Results (copies/mL) | Mean (log 10 copies/mL) | SD (log 10 copies/mL) | CV (%) | ||
| Run 1 | Run 2 | Run 3 | |||||
| L0 | 7.76 × 107 - 6.17 × 108 | 1.65 × 108 | 1.84 × 108 | 1.52 × 108 | 8.22 | 0.04 | 0.5 |
| L1 | 1.48 × 107 - 1.18 × 108 | 9.21 × 107 | 1.01 × 108 | 9.23 × 107 | 7.98 | 0.02 | 0.3 |
| L2 | 1.59 × 106 - 1.26 × 107 | 8.54 × 106 | 9.00 × 106 | 9.20 × 106 | 6.95 | 0.02 | 0.3 |
| L3 | 1.66 × 105 - 1.32 × 106 | 7.06 × 105 | 6.13 × 105 | 9.55 × 105 | 5.87 | 0.10 | 1.7 |
| L4 | 1.82 × 104 - 1.48 × 105 | 8.60 × 104 | 3.84 × 104 | 6.30 × 104 | 4.77 | 0.18 | 3.8 |
| L5 | 1.51 × 103 - 1.23 × 104 | 1.07 × 104 | 6.20 × 103 | 8.82 × 103 | 3.92 | 0.12 | 3.1 |
| L6 | < 1000 | 524 | 442 | 581 | 2.71 | 0.06 | 2.2 |
For evaluation of the intra-assay variation of the Fosun real-time PCR kits, three samples with high, medium and low HBV titers were tested. Each sample was tested in 12 replicates. Table 2 shows the mean log 10 (copies/mL) viral loads, SD and coefficient of variation (CV) of the samples. The viral load values from the Fosun real-time PCR assay were highly reproducible over the various HBV titers (CV, 1.4%-8.1%), especially for the samples with high and medium HBV titers, and the intra-assay CV was 1.4% and 2%, respectively.
| No. of samples | Replicates | Mean of HBV DNA | SD | CV (%) |
| level (log10 copies/mL) | ||||
| 3768 | 12 | 3.97 | 0.32 | 8.1 |
| 3785 | 12 | 6.00 | 0.12 | 2.0 |
| 3770 | 12 | 7.84 | 0.11 | 1.4 |
A total of 158 clinical serum samples were tested in parallel with the Fosun real-time PCR kits and the COBAS Amplicor HBV Monitor system. Samples were divided into three groups: group A, samples from healthy blood donors (n = 40); group B, samples from HBV infected patients with viral loads within the range of both tests (n = 43); group C, samples from HBV infected patients with viral loads above the upper limit of the Amplicor test (n = 75). These samples were diluted and re-tested with the Amplicor test. Viral loads were converted into logarithms. In group A, all the samples were negative for HBV with both assays. A comparison between the two assays was performed from samples in group B and C. Spearman’s coefficient showed a significant correlation between the two assays (r = 0.948, P < 0.0001, Figure 1). The differences between viral loads obtained with the two assays versus the average for each specimen was analyzed with the Bland-Altman method[8] (Figure 2). Of the 158 samples, two (1.3%) samples had viral loads slightly above the upper limit and one (0.6%) sample was below the lower limit of the Fosun real-time PCR. However, viral loads of 75 (47.5%) samples were above the upper limit of the Amplicor test. In addition, HBV genotypes did not influence the quantitative results obtained with both assays.
The assay times and costs of both tests were compared. For a 24-sample work unit (controls were handled in the same way as clinical samples), Fosun real-time PCR required 3 h and the Amplicor test required 7 h. The cost of the Fosun real-time PCR per test, including consumables and DNA extraction reagents, was 5 US dollars. The cost of the Amplicor test was approximately 100 US dollars per test. If the samples were diluted and retested, the cost would be much higher.
China has the greatest burden of HBV infection in the world. Many commercially available HBV DNA assays in the United States and Europe are unaffordable for Chinese patients. Reliable but inexpensive HBV DNA assays are very important for control and treatment of HBV infection in low-income countries such as China. Fosun real-time PCR HBV DNA kits are produced by a Chinese company and approved by the Chinese SFDA for in vitro diagnostic use. Compared to the COBAS Amplicor test, this assay has equivalent reliability at a reduced cost.
Many studies have shown that real-time PCR is accurate, reproducible, sensitive, and rapid for quantitative detection of HBV levels in clinical samples[131417–21]. Real-time PCR also has the widest dynamic range among all the assays available commercially[14]. In this study, Fosun real-time PCR was able to quantitatively detect HBV in all seven reference sera from NICPBP, which showed a dynamic range of 8 logs. The majority (97.5%) of the HBV-positive samples used in this study were quantified within the manufacturer’s recommended dynamic range for the Fosun real-time PCR, which indicates that this assay is superior to the Amplicor test for detection of HBV DNA in patients with high viral loads.
The results for the 158 clinical samples obtained with the Fosun real-time PCR and Amplicor tests were concordant very well. Comparison of the two assays for quantitative detection of HBV DNA levels in the 118 serum samples showed that the two sets of results were highly correlated (r = 0.948, P < 0.001). This was in accordance with the results of previous studies that have evaluated other real-time PCR assays for quantitation of HBV DNA[131422–26].
One of the advantages of the Fosun real-time PCR kits is its low cost compared to that of the Amplicor test (5 versus 100 US dollars per test) and other commercially available tests[14]. With its low cost, it can be used to monitor antiviral therapy in economically disadvantaged patients with chronic HBV infection. As recommended by the American Association for Study of Liver Diseases and the Asian-Pacific Consensus Update Working Party on Chronic Hepatitis B, HBV DNA should be monitored at least every 3 mo during therapy. After the end of therapy, HBV DNA should be monitored monthly for the first 3 mo and then every 3-6 mo[2728]. This will be a great economic burden for chronic hepatitis B patients in countries such as China, where most people have no medical insurance. Reliable and inexpensive assays for quantitation of HBV DNA are helpful for controlling HBV infection in such countries.
As with many other real-time PCR tests, the Fosun real-time PCR has no internal control included in the test. Internal controls can compensate for the differences in DNA extraction efficiency between specimens, and possible PCR inhibition in the reaction mixture[1429–31]. In addition, standardization with international reference standards is also needed for wide acceptance of this assay. Further comparison with other commercially available assays such as the COBAS TaqMan 48 real-time PCR system will be helpful for evaluating the clinical performance of the Fosun real-time PCR assay.
In conclusion, the Fosun real-time PCR HBV DNA kit, with its much lower costs, is capable of providing reliable and rapid HBV DNA quantitation, which is useful for monitoring HBV DNA levels in patients with chronic hepatitis B.
Commercial real-time PCR assays are widely used for quantitation of hepatitis B virus (HBV) DNA in China. However, these assays have not been fully evaluated for their clinical performance.
Several methods are routinely used for quantitation of HBV DNA in serum or plasma samples. However, differences do exist among these methods. Comparison and evaluation of these methods are the focus of research efforts.
Lindh et al and Gordillo et al have evaluated the clinical performance of commercial real-time PCR assays. However, they have not compared the cost and time between the real-time PCR and reference methods.
This study compared a real-time PCR assay widely used in China with the COBAS Amplicor test, which is widely used in the United States and Europe, for detection of HBV DNA in serum samples. Clinical performance, time required, and cost of the assays were analyzed.
This commercial PCR assay is suitable for quantitation of HBV DNA in serum samples.
TaqMan PCR is one of the real-time PCR methods for the determination of copy number of PCR templates. It requires a pair of PCR primers and a fluorogenic probe, which is an oligonucleotide with both a reporter fluorescent dye and a quencher dye attached. The fluorescence can be monitored at frequent intervals during the PCR reaction with a real-time PCR machine.
This is a well designed and prepared study with important conclusions. The most important point is that the real-time PCR is significantly cheaper than other commercial tests.
| 2. | Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733-1745. |
| 3. | Liaw YF, Tai DI, Chu CM, Chen TJ. The development of cirrhosis in patients with chronic type B hepatitis: a prospective study. Hepatology. 1988;8:493-496. |
| 4. | Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942-1956. |
| 5. | Yeo W, Mo FK, Chan SL, Leung NW, Hui P, Lam WY, Mok TS, Lam KC, Ho WM, Koh J. Hepatitis B viral load predicts survival of HCC patients undergoing systemic chemotherapy. Hepatology. 2007;45:1382-1389. |
| 6. | Yim HJ, Byun KS, Chang YJ, Suh YS, Yeon JE, Lee CH, Kwon JA, Yoo W, Kim SO, Hong SP. Levels of hepatitis B virus (HBV) replication during the nonreplicative phase: HBV quantification by real-time PCR in Korea. Dig Dis Sci. 2007;52:2403-2409. |
| 7. | Liu CJ, Chen PJ, Lai MY, Lin FY, Wang T, Kao JH, Chen DS. Viral factors correlate with hepatitis B e antigen seroconverson in patients with chronic hepatitis B. Liver Int. 2006;26:949-955. |
| 8. | Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, Chen DS. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193:1258-1265. |
| 9. | Chan HL, Tsang SW, Liew CT, Tse CH, Wong ML, Ching JY, Leung NW, Tam JS, Sung JJ. Viral genotype and hepatitis B virus DNA levels are correlated with histological liver damage in HBeAg-negative chronic hepatitis B virus infection. Am J Gastroenterol. 2002;97:406-412. |
| 10. | Yao JD, Beld MG, Oon LL, Sherlock CH, Germer J, Menting S, Se Thoe SY, Merrick L, Ziermann R, Surtihadi J. Multicenter evaluation of the VERSANT hepatitis B virus DNA 3.0 assay. J Clin Microbiol. 2004;42:800-806. |
| 11. | Yuan HJ, Yuen MF, Wong DK, Sum SS, Lai CL. Clinical evaluation of the digene hybrid capture II test and the COBAS AMPLICOR monitor test for determination of hepatitis B virus DNA levels. J Clin Microbiol. 2004;42:3513-3517. |
| 12. | Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M, Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899-2903. |
| 13. | Ronsin C, Pillet A, Bali C, Denoyel GA. Evaluation of the COBAS AmpliPrep-total nucleic acid isolation-COBAS TaqMan hepatitis B virus (HBV) quantitative test and comparison to the VERSANT HBV DNA 3.0 assay. J Clin Microbiol. 2006;44:1390-1399. |
| 14. | Sum SS, Wong DK, Yuen JC, Lai CL, Yuen MF. Comparison of the COBAS TaqMan HBV test with the COBAS Amplicor monitor test for measurement of hepatitis B virus DNA in serum. J Med Virol. 2005;77:486-490. |
| 15. | Pas SD, Fries E, De Man RA, Osterhaus AD, Niesters HG. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J Clin Microbiol. 2000;38:2897-2901. |
| 16. | Bland JM, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085-1087. |
| 17. | Chen RW, Piiparinen H, Seppanen M, Koskela P, Sarna S, Lappalainen M. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J Med Virol. 2001;65:250-256. |
| 18. | Lindh M, Hannoun C. Dynamic range and reproducibility of hepatitis B virus (HBV) DNA detection and quantification by Cobas Taqman HBV, a real-time semiautomated assay. J Clin Microbiol. 2005;43:4251-4254. |
| 19. | Lu YQ, Han JX, Qi P, Xu W, Zu YH, Zhu B. Rapid quantification of hepatitis B virus DNA by real-time PCR using efficient TaqMan probe and extraction of virus DNA. World J Gastroenterol. 2006;12:7365-7370. |
| 20. | Liu Y, Hussain M, Wong S, Fung SK, Yim HJ, Lok AS. A genotype-independent real-time PCR assay for quantification of hepatitis B virus DNA. J Clin Microbiol. 2007;45:553-538. |
| 21. | Olioso D, Boaretti M, Ligozzi M, Lo Cascio G, Fontana R. Detection and quantification of hepatitis B virus DNA by SYBR green real-time polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 2007;26:43-50. |
| 22. | Gordillo RM, Gutierrez J, Casal M. Evaluation of the COBAS TaqMan 48 real-time PCR system for quantitation of hepatitis B virus DNA. J Clin Microbiol. 2005;43:3504-3507. |
| 23. | Weiss J, Wu H, Farrenkopf B, Schultz T, Song G, Shah S, Siegel J. Real time TaqMan PCR detection and quantitation of HBV genotypes A-G with the use of an internal quantitation standard. J Clin Virol. 2004;30:86-93. |
| 24. | Welzel TM, Miley WJ, Parks TL, Goedert JJ, Whitby D, Ortiz-Conde BA. Real-time PCR assay for detection and quantification of hepatitis B virus genotypes A to G. J Clin Microbiol. 2006;44:3325-3333. |
| 25. | Lole KS, Arankalle VA. Quantitation of hepatitis B virus DNA by real-time PCR using internal amplification control and dual TaqMan MGB probes. J Virol Methods. 2006;135:83-90. |
| 26. | Hochberger S, Althof D, Gallegos de Schrott R, Nachbaur N, Rock H, Leying H. Fully automated quantitation of hepatitis B virus (HBV) DNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol. 2006;35:373-380. |
| 27. | Liaw YF, Leung N, Guan R, Lau GK, Merican I, McCaughan G, Gane E, Kao JH, Omata M. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2005 update. Liver Int. 2005;25:472-489. |
| 28. | Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507-539. |
| 29. | Garson JA, Grant PR, Ayliffe U, Ferns RB, Tedder RS. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods. 2005;126:207-213. |
| 30. | Stocher M, Leb V, Berg J. A convenient approach to the generation of multiple internal control DNA for a panel of real-time PCR assays. J Virol Methods. 2003;108:1-8. |