Published online Jun 28, 2008. doi: 10.3748/wjg.14.3849
Revised: May 19, 2008
Accepted: May 26, 2008
Published online: June 28, 2008
AIM: To observe the inhibition of hepatitis B virus (HBV) replication and expression in HepG2.2.15 cells by combination of small interfering RNAs (siRNAs).
METHODS: Recombinant plasmid psil-HBV was constructed and transfected into HepG2.2.15 cells. At 48 h, 72 h and 96 h after transfection, culture media were collected and cells were harvested for HBV replication assay. HBsAg and HBeAg in the cell culture medium were detected by enzyme-linked immunoadsorbent assay (ELISA). Intracellular viral DNA and covalently closed circular DNA (cccDNA) were quantified by real-time polymerase chain reaction (PCR). HBV viral mRNA was reverse transcribed and quantified by reverse-transcript PCR (RT-PCR).
RESULTS: siRNAs showed marked anti-HBV effects. siRNAs could specifically inhibit the expression of HBsAg and the replication of HBV DNA in a dose-dependent manner. Furthermore, combination of siRNAs, compared with individual use of each siRNA, exerted a stronger inhibition on antigen expression and viral replication. More importantly, combination of siRNAs significantly suppressed HBV cccDNA amplification.
CONCLUSION: Combination of siRNAs mediates a stronger inhibition on viral replication and antigen expression in HepG2.2.15 cells, especially on cccDNA amplification.
- Citation: Xin XM, Li GQ, Jin YY, Zhuang M, Li D. Combination of small interfering RNAs mediates greater suppression on hepatitis B virus cccDNA in HepG2.2.15 cells. World J Gastroenterol 2008; 14(24): 3849-3854
- URL: https://www.wjgnet.com/1007-9327/full/v14/i24/3849.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.3849
Hepatitis B virus (HBV) is a major cause for acute and chronic hepatitis in humans. Although recombinant vaccines are widely available, about 400 million people have chronic HBV infection worldwide. Chronic infection may also have serious consequences, and nearly 25% people with chronic HBV infection would die due to untreatable liver cancer[1]. The deaths of liver cancer patients resulting from chronic HBV infection exceeds one million per year worldwide[2]. Therefore, it is important to develop effective strategies for the treatment of HBV-infected patients.
Nucleotide analogues, such as lamivudine, can effectively inhibit HBV DNA synthesis[34] and are widely used in the treatment of HBV-infected patients. However, analysis of viral kinetics during lamivudine therapy revealed that a prolonged treatment is required since lamivudine does not completely inhibit viral replication and the rate of clearance for infected hepatocytes is slow[5]. HBV is not a cytopathogenic virus and hepatocytes are normally long-lived and their half-life is estimated to be 6-12 mo or longer, explaining the requirement for a long-term antiviral treatment course, which is associated with the selection of drug-resistant mutants[6].
During HBV replication, viral covalently closed circular DNA (cccDNA) serves as the template for viral transcription and its production is regulated and amplified by an intra-cellular pathway in which newly synthesized genomic DNA is recycled to the nuclei[78]. This process establishes a steady pool of nuclear cccDNA, which is maintained during the life of infected hepatocytes. It is likely that cccDNA may reactivate synthesis of viral transcript and protein, leading to a rebound of active viral replication. Thus, elimination of cccDNA from infected hepatocytes still remains a challenge in therapy for HBV-infected patients. A potent agent used in anti-HBV therapy should be evaluated with special emphasis on its inhibitory effect against the amplification of cccDNA.
Small interfering RNAs (siRNA) are double-stranded RNA molecules, approximately 21 nucleotides in length that hybridize to a homologous mRNA target and result in degradation of mRNA[9]. This posttranscriptional gene silencing process, called RNA interference (RNAi), is evolutionarily conserved in both plants and eukaryotic cells[1011]. Due to the high sequence specificity and efficiency, RNAi can be used in functional genomic studies such as human immunodeficiency virus (HIV), hepatitis C virus (HCV), and influenza virus, etc[12–14]. HBV is one of the major candidates for RNAi, as its pregenomic RNA is a key intermediate for maintaining DNA replication via reverse transcription in the viral life cycle. Many studies have demonstrated that siRNA targeting different regions of the HBV genome can block viral replication and antigen expression[15–19]. Our previous study showed that siRNA targeting HBV nuclear localization signal (NLS) can inhibit viral DNA synthesis and cccDNA amplification[20]. As HBV infection is a cellular infection involving multiple genes, we hypothesize that combination of siRNAs targeting different sequences along the HBV NLS mediates a greater inhibitory efficacy on cccDNA amplication. If this is the case, it would be of significance in the treatment of HBV infection. In this study, we analyzed the kinetics of HBV genome replication during siRNA therapy and tested the antiviral capacity of combination therapy in comparison to monotherapy in HepG2.2.15 cells, showing that combination of siRNAs exhibits better effects on the inhibition of HBV replication, especially at viral cccDNA level.
Dulbecco’s modified Eagle’s medium (DMEM) and G418 were purchased from GIBCO BRL (USA). HepG2.2.15 cells were maintained in our laboratory. Plasmid psi/U6 was supplied by Wuhan JS Biotech (China). All polymerase chain reaction (PCR) primers were synthesized by Shanghai Boya Biological Company (China). Trizol, M-MLV reverse transcriptase, Lipofection 2000 reagent were purchased from Invitrogen Company (USA). Restriction enzymes were purchased from New England Bio-laboratory (Beijing, China).
siRNA expression plasmid was generated as previously described[21]. Briefly, 21-nucleotide long inverted sequences were cloned into the plasmid pGenesil/U6. Five thymidines were inserted into the downstream anti-sense strand to provide a stop signal for the polymerase III RNA polymerase. The sense strand of hairpin was homologous to the target mRNA (NLS region) as analyzed by BLAST in the NCBI database. Oligonucletides used to code for the sense strand of siRNA included siRNA1 (5’-AAG ATCTCAATCTCGGGAATC-3’), siRNA2 (5’-CAGGTCCCCTAGAAGAAGAAC-3’), and control siRNAHK (5’-ACTACCGTTGTTATAGGTG-3’). All the plasmids constructed were confirmed by endonuclease digestion and DNA sequencing.
The human hepatoma cell line, HepG2.2.15, was maintained in Dubecco’s modified Eagle’s medium (DMEM) with 10% fatal bovine serum and 200 &mgr;g/mL G418 in 5% CO2-humidified air as previously described[22]. Cells were cultur at a density of 3 × 105 cells per well in 6-well plates. Twenty-four hours after incubation, the cells were transfected with 4 &mgr;g siRNA-expressed plasmid using the Lipofection 2000 reagent. We removed the medium, washed the cells with warmed PBS, and added fresh medium every 24 h. At 48, 72 and 96 h after transfection, culture media were collected and cells were harvested for HBV replication assay. All experiments were performed in triplicate and divided into five groups.
To assess the effects of RNAi on viral antigen expression, HBsAg and HBeAg levels in culture medium were measured with an enzyme-linked immunosorbent assay (ELISA) kit following its manufacture’s instructions.
Antiviral activities were evaluated by reverse-transcript PCR (RT-PCR). Total RNA was extracted directly from the transfected cells using Trizol reagent. Then, RNA was denatured for 5 min at 70°C, immediately cooled in ice water, reverse transcribed using M-MLV reverse transcriptase. A RT-PCR experiment targeting β-actin gene was run as an internal control. The primer sequences used are HBV forward (5’-ACCTCTGCCTAATCATCTC-3’) and reverse (5’-GTAAGACAGGAAATGTGAAAC-3’), β-actin forward (5’-GTCGGTGTGAACGGATTT-3’) and reverse (5’-ACTCCACGACGTACTCAGC-3’). The PCR amplification product was analyzed by 1.2% agarose gel electrophoresis.
Real-time PCR was performed to quantify the HBV DNA or cccDNA using a HBV fluorescence detection kit following its manufacturer’s protocol. For measurement of viral DNA, DNA was extracted from the culture supernatant using a QIAamp DNA mini kit. cccDNA was isolated from the transfection cells to quantify its level and examined every 24 h post-transfection according to its manufacture’s protocol. Reactions with no reverse transcriptase enzyme added were performed in parallel. The inhibition ratio of HBV DNA was calculated according to the formula: 1-log (treated sample fluorescent intensity)/log (control fluorescent intensity) × 100%.
To evaluate the dose-dependent effects of siRNA on HBV gene expression, a series of experiments were conducted in HepG2.2.15 cells transfected with different plasmids at the concentration of 2 &mgr;g, 3 &mgr;g and 4 &mgr;g, respectively. The transfection cells were harvested and cell culture supernatant was collected 96 h post-transfection for further examination. Several parameters of HBV were measured including HBsAg, HBV DNA as described above.
Statistical analysis was performed using the SPSS 12.0 software. The results were expressed as mean ± SD and compared using F-test and one-way ANOVA. P < 0.05 was considered statistically significant.
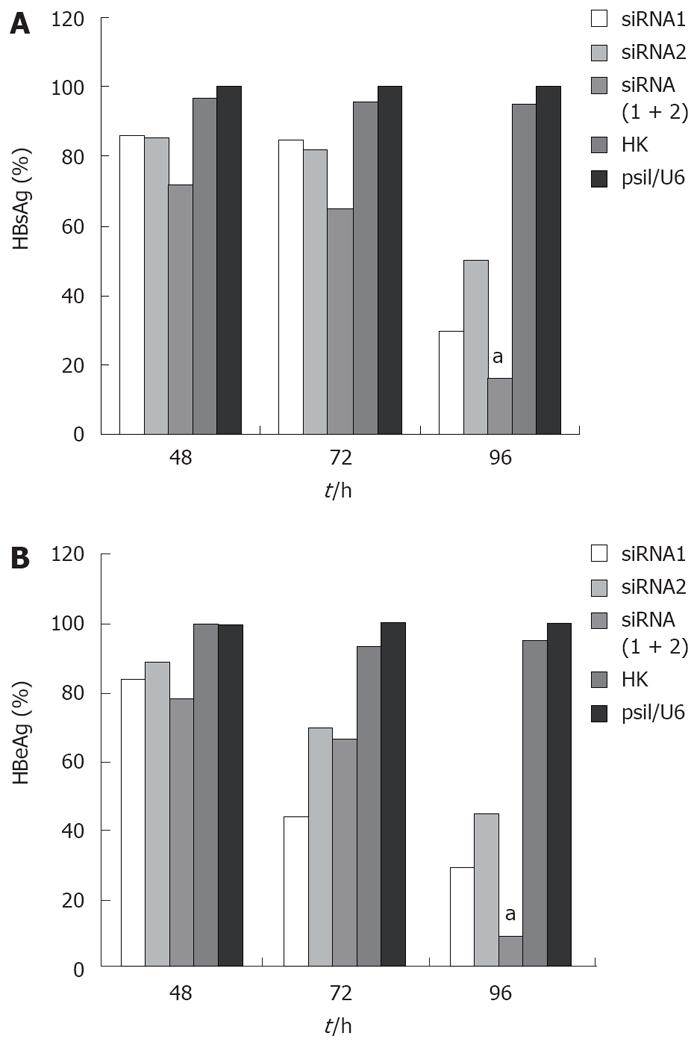
To test whether siRNA could effectively inhibit the expression of viral proteins in HepG2.2.15 cells, HBsAg and HBeAg in culture media were detected by ELISA. For HBsAg, cells transfected with the control siRNA produced an equal secretion with those treated with empty vector (P < 0.05) as expected in the absence of any specific silencing response, whereas cells expressing HBV siRNA gave a very different result. The HBsAg level in all the cells integrated with three siRNAs was significantly reduced compared with those transfected with empty vector, and the greatest reduction rate was 83.89% in the combined therapy group 96 h post-transfection (Figure 1A). HBeAg was also reduced in the selected siRNA-transfected cells, and the greatest reduction rate was 91.07% in the combined therapy group 96 h post-transfection (Figure 1B). For both HBsAg and HBeAg, combination therapy for siRNAs was more potent than any individual therapy.
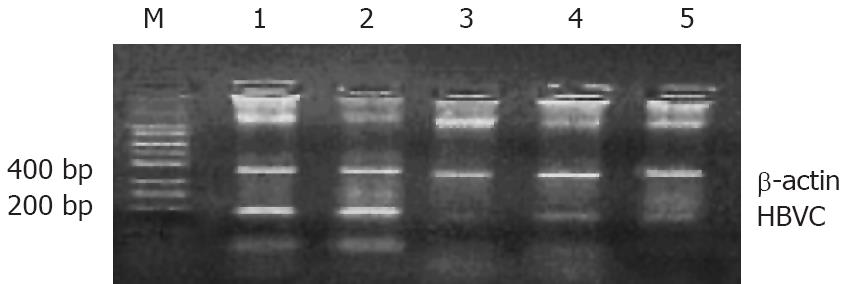
To determine whether viral mRNA is efficiently degraded by siRNA, RT-PCR was performed 48 h, 72 h and 96 h after transfection. The results demonstrated that the mRNA level of each tested siRNA was markedly reduced, whereas the empty plasmid had no effect on mRNA level (Figure 2). Combination therapy with all the siRNAs produced a stronger inhibition on viral transcripts compared with the therapy with one siRNA. The HepG2.2.15 cells treated with combined siRNAs for 96 h did not contain any detectable mRNA, whereas mRNA was abundant in cells treated with empty vector.
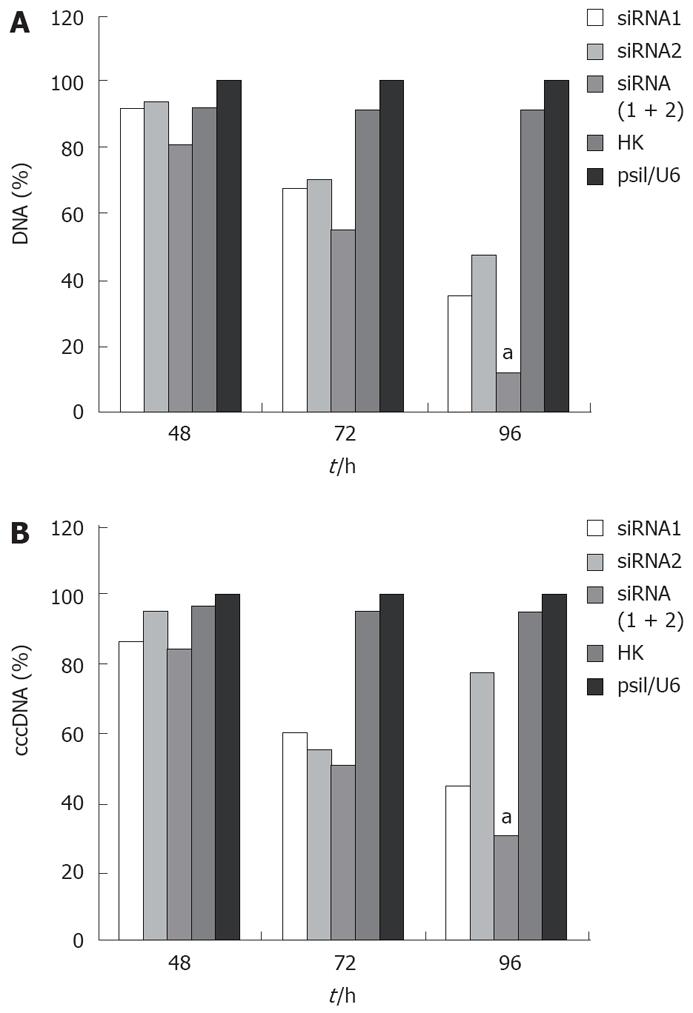
The effect of siRNA silencing on HBV DNA was investigated by quantitative real-time PCR, which can detect 104-108 copies of HBV DNA. The level of viral DNA in cells transfected with combined siRNAs was significantly lower than that in cells treated with empty vector. Treatment with irrelevant control siRNA slightly decreased the level of viral DNA in cells transfected with siRNA (Figure 3A). Furthermore, combined siRNA1 and siRNA2 showed a greater inhibitory effect (88.6%) on viral DNA replication than siRNA1 or siRNA2 alone (P < 0.05).
HBV cccDNA is an important parameter in the therapy for chronic HBV infection. To evaluate if combination of siRNAs can effectively inhibit viral cccDNA, we isolated viral cccDNA from the transfected cells every 24 h post-trasfection. Quantitative assay revealed that HBV cccDNA levels were decreased by 50.4%, 22.31%, and 69.83% in the cells transfected with siRNA1, siRNA2 and siRNA (1 + 2), respectively, compared to the level of cells treated with empty plasmid 96 h post-transfection. Meanwhile, HBV cccDNA levels did not significantly change in cells treated with control siRNA and empty vector. As expected, the use of siRNA1 and siRNA2 synergistically suppressed the viral cccDNA activities compared with siRNA1 or siRNA2 alone (P < 0.05, Figure 3B). These results were reproducibly observed in three independent experiments.
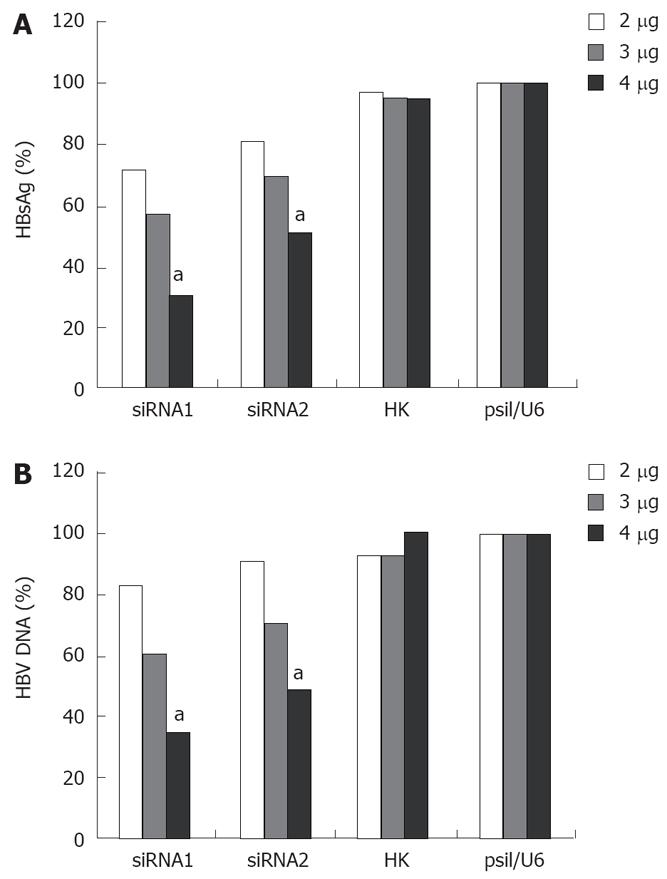
As shown in Figure 4, siRNA had an obvious dose-dependent effect on HBV replication and antigen expression 96 h post-transfection. When the cells were transfected with a low concentration of siRNA, siRNA had almost no effect on HBV replication and antigen expression. However, a more significant inhibition of HBsAg and DNA in the cells treated at the concentration of 4 &mgr;g was observed. The level of HBsAg was decreased by 29%, 43% or 70% after siRNA1 treatment, and 9%, 30% or 50% after siRNA2 treatment, respectively, at the concentrations of 2 &mgr;g, 3 &mgr;g, 4 &mgr;g (Figure 4A). The level of HBV DNA was reduced by 17%, 39% or 65% after siRNA1 treatment, and 9%, 29% or 51% after siRNA2 treatment, respectively, at the concentrations of 2 &mgr;g, 3 &mgr;g, 4 &mgr;g (Figure 4B).
Chronic HBV infection is one of the most serious diseases threatening human health. RNAi technology provides an alternative strategy to combat HBV infection[2324]. It was reported that viral mutations could escape single synthetic siRNA treatments, such as HBV, HCV and HIV[25–27]. One strategy to address the problem is to generate multiple siRNA molecules that can target different sites on the viral genome. The other is to choose target sequences in the relatively conserved region. Recently, combination therapy has emerged as a new approach to the treatment of chronic HBV infection with the objective to decrease the viral load to the lowest possible levels[2829]. This is then followed by a continued chemotherapy with another nucleotide analog or IFN-α in order to eliminate the remained viral load. Following the same strategy, we also investigated the effect of combination of siRNAs in comparison to monotherapy in HepG2.2.15 cells. As HBV infection is a pathological process involving multiple genes, it will be ideal to develop a combination strategy for siRNAs that can knock down the expression of multiple pathogenic viral antigens as well as inhibit viral replication. During the progress of combination therapy, several siRNAs may block multiple sites in viral genome and make it difficult to repair immediately, thus achieving greater suppression on HBV infection than a single siRNA.
The HepG2.2.15 cell line, a derivative of human HepG2 hepatoma cells, has been used as an in vitro stable HBV-producing model[22]. The cell line can be transformed with a head-tail dimmer of HBV DNA. Using this method, we could simulate the natural condition in which cells can still stably produce mRNA, antigen and viral particles. Specific siRNA targeting different sites on HBV NLS region were transfected and monitored in HepG2.2.15cells. As a result, the load of HBsAg, HBeAg, and mRNA was inhibited to some extent. Meanwhile, real-time PCR revealed that the number of viral DNA and cccDNA copies was markedly decreased. This inhibition was highly selective, sequence-specific and dose-dependent since control siRNA had almost no inhibitory effect on the expression or replication of HBV.
The selected siRNAs showed marked anti-HBV effects. Surprisingly, combination of siRNAs, compared with a single siRNA, exerted a stronger inhibition on antigen expression and viral replication, even though the final concentration of siRNA used in the therapy was the same. More importantly, combination therapy significantly suppressed HBV cccDNA amplification.
In summary, combination of siRNAs targeting various regions of HBV NLS inhibits viral replication and suppresses cccDNA amplication and can be used in the treatment of HBV infection.
Hepatitis B virus (HBV) is a major cause for acute and chronic hepatitis in humans. The development of an effective therapy for HBV infection is still a challenge. As HBV infection is a cellular infection involving multiple genes, progress in RNA interference (RNAi) has shed slight on developing a new anti-HBV strategy.
siRNA targeting HBV nuclear localization signal (NLS) can inhibit viral DNA synthesis and cccDNA amplification. In this study, the authors hypothesize that combination of siRNAs targeting different sequences along HBV NLS mediates greater inhibitory effects on cccDNA. If this is the case, it would be of significance in the treatment of HBV infection.
In this study, the selected siRNA showed marked anti-HBV effects. Surprisingly, combination of siRNAs, compared with a single siRNA, exerted a stronger inhibition on antigen expression and viral replication, even though the final concentration of siRNA used in the therapy was the same. More importantly, combination therapy significantly suppressed HBV cccDNA amplification.
Combination of siRNAs targeting various regions of HBV NLS not only inhibits viral replication, but also suppresses cccDNA amplification, indicating that it can be used in the treatment of HBV infection.
The effect of combination of siRNAs on suppressing HBsAg and HBeAg expression, and levels of HBV mRNA, DNA and cccDNA in vitro was studied in this study. This manuscript provides some important information about the treatment of chronic HBV infection. It is very interesting paper and has certain value.
| 1. | Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395-403. |
| 2. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. |
| 3. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. |
| 4. | Papatheodoridis GV, Dimou E, Papadimitropoulos V. Nucleoside analogues for chronic hepatitis B: antiviral efficacy and viral resistance. Am J Gastroenterol. 2002;97:1618-1628. |
| 5. | Ayres A, Bartholomeusz A, Lau G, Lam KC, Lee JY, Locarnini S. Lamivudine and Famciclovir resistant hepatitis B virus associated with fatal hepatic failure. J Clin Virol. 2003;27:111-116. |
| 6. | Lau DT, Khokhar MF, Doo E, Ghany MG, Herion D, Park Y, Kleiner DE, Schmid P, Condreay LD, Gauthier J. Long-term therapy of chronic hepatitis B with lamivudine. Hepatology. 2000;32:828-834. |
| 7. | Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451-460. |
| 8. | Rollier C, Sunyach C, Barraud L, Madani N, Jamard C, Trepo C, Cova L. Protective and therapeutic effect of DNA-based immunization against hepadnavirus large envelope protein. Gastroenterology. 1999;116:658-665. |
| 9. | Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550-553. |
| 10. | Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985-4990. |
| 12. | Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435-438. |
| 13. | Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014-2018. |
| 14. | Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430-434. |
| 15. | Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047-6052. |
| 16. | Tang N, Huang AL, Zhang BQ, Yan G, He TC. [Potent and specific inhibition of hepatitis B virus antigen expression by RNA interference]. Zhonghua Yixue Zazhi. 2003;83:1309-1312. |
| 17. | Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773-778. |
| 18. | Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764-770. |
| 19. | Wu KL, Zhang X, Zhang J, Yang Y, Mu YX, Liu M, Lu L, Li Y, Zhu Y, Wu J. Inhibition of Hepatitis B virus gene expression by single and dual small interfering RNA treatment. Virus Res. 2005;112:100-107. |
| 20. | Li GQ, Gu HX, Li D, Xu WZ. Inhibition of Hepatitis B virus cccDNA replication by siRNA. Biochem Biophys Res Commun. 2007;355:404-408. |
| 21. | Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515-5520. |
| 22. | Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005-1009. |
| 23. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. |
| 24. | Zhang XN, Xiong W, Wang JD, Hu YW, Xiang L, Yuan ZH. siRNA-mediated inhibition of HBV replication and expression. World J Gastroenterol. 2004;10:2967-2971. |
| 25. | Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601-2605. |
| 26. | Konishi M, Wu CH, Kaito M, Hayashi K, Watanabe S, Adachi Y, Wu GY. siRNA-resistance in treated HCV replicon cells is correlated with the development of specific HCV mutations. J Viral Hepat. 2006;13:756-761. |
| 27. | Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708-716. |
| 28. | Li GQ, Xu WZ, Wang JX, Deng WW, Li D, Gu HX. Combination of small interfering RNA and lamivudine on inhibition of human B virus replication in HepG2.2.15 cells. World J Gastroenterol. 2007;13:2324-2327. |
| 29. | Colledge D, Civitico G, Locarnini S, Shaw T. In vitro antihepadnaviral activities of combinations of penciclovir, lamivudine, and adefovir. Antimicrob Agents Chemother. 2000;44:551-560. |