Published online Jun 21, 2008. doi: 10.3748/wjg.14.3733
Revised: April 23, 2008
Accepted: April 30, 2008
Published online: June 21, 2008
AIM: To construct eukaryotic expression plasmids of full-length Hepatitis B Virus (HBV) genotype C genome, which contain lamivudine-resistant mutants (YIDD, YVDD) or wild-type strain (YMDD), and to observe the expression of HBV DNA and antigens [hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg)] of the recombinant plasmids in HepG2 cells.
METHODS: Three HBV full-length genomes were amplified from the plasmids pMD18T-HBV/YIDD, pMD18T-HBV/YVDD and pMD18T-HBV/YMDD, using PCR. Three recombinant plasmids were generated by inserting each of the PCR products into the eukaryotic expression vector pcDNA3.1 (+), between the EcoRI and HindIII sites. After being characterized by restriction endonuclease digestion, and DNA sequence analysis, the recombinant plasmids were transfected into HepG2 cells. At 48 and 72 h post-transfection, the levels of intracellular viral DNA replication were detected by real-time PCR, and the expression of HBsAg and HBeAg in the cell culture supernatant was determined by ELISA.
RESULTS: Restriction endonuclease digestion and DNA sequence analysis confirmed that the three recombinant plasmids were correctly constructed. After transfecting the plasmids into HepG2 cells, high levels of intracellular viral DNA replication were observed, and HBsAg and HBeAg were secreted into the cell culture supernatant.
CONCLUSION: Eukaryotic expression plasmids pcDNA3.1 (+)-HBV/YIDD, pcDNA3.1 (+)-HBV/YVDD or pcDNA3.1 (+)-HBV/YMDD, which contained HBV genotype C full-length genome, were successfully constructed. After transfection into HepG2 cells, the recombinant plasmids efficiently expressed HBV DNA, HBsAg and HBeAg. Our results provide an experimental basis for the further study of HBV lamivudine-resistant mutants.
- Citation: Xu WZ, Fang Y, Li D, Wang Y, Shang QL, Li GQ, Teng X, Gu HX. Construction and expression of eukaryotic plasmids containing lamivudine-resistant or wild-type strains of Hepatitis B Virus genotype C. World J Gastroenterol 2008; 14(23): 3733-3738
- URL: https://www.wjgnet.com/1007-9327/full/v14/i23/3733.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.3733
Hepatitis B Virus (HBV) infection has become one of the most serious health problems worldwide. In China, HBV prevalence is especially high. Presently, eight genotypes of HBV (A-H) have been identified, based on divergence over the entire genomic sequence of ≥ 8%[1–4]. Different HBV genotypes have specific geographical distributions[25]. According to previous studies, genotypes B and C are predominant in China[67]. In Heilongjiang Province in northern China, HBV genotype C is dominant[89].
Lamivudine, a potent, non-toxic inhibitor of HBV replication in chronically infected patients, is currently one of the most effective anti-HBV drugs in the clinic. Unfortunately, it has been found that long-term use of lamivudine leads to emergence of HBV YMDD mutants, which has been demonstrated to be associated with lamivudine resistance[1011]. In YMDD variants, the methionine of the YMDD motif in HBV polymerase is substituted with either isoleucine, designated as YIDD, or valine, designated as YVDD. Much clinical data has indicated that patients who have developed HBV YMDD mutations show deterioration of their physical condition, and rebound of virus load in their serum[12–14]. In this study, we constructed the eukaryotic expression plasmids of HBV genotype C full-length genome, which contained wild-type, YVDD mutation or YIDD mutation, respectively. All these recombinant plasmids were shown to be able to express HBV DNA and antigens in vitro.
Platinum Pfx DNA polymerase, T4 DNA ligase, and Lipofection 2000 reagent were purchased from Invitrogen (Carlsbad, CA, USA). Restriction endonucleases EcoRI and HindIII were purchased from New England Biolabs (Beijing, China). Dulbecco’s modified Eagle’s medium (DMEM) was purchased from GIBCO BRL (Gaithersburg, MD, USA). PCR primers were synthesized by Shanghai Sangon Biological Engineering, Technology and Services (Shanghai, China). DNA sequencing was performed by Invitrogen (Beijing, China). Enzyme immunoassay kit was purchased from Shanghai Kehua Biochemical Laboratory (Shanghai, China). Quantitative HBV PCR Fluorogence Diagnostic Kit was purchased from PG Biotechnology (Nanjing, China). Axygen DNA Mini kit was purchased from Axygen Biosciences (Union City, CA, USA). The recombinant plasmids pMD18T-HBV were constructed in our laboratory. The HBV genotype C full-length genome of wild-type strain or YIDD, YVDD mutants were obtained from serum of chronic HBV patients, and inserted into the vector pMD18T. The expression vector pcDNA3.1 (+), HepG2 cells and Escherichia coli DH5α were maintained in our laboratory.
The HBV full-length genome of wild-type HBV DNA, YVDD or YIDD mutants was amplified from the plasmids pMD18T-HBV, by PCR using a primer set that consisted of a sense primer: 5'TACCATGGCCCTTTTTCACCTCTGCCTAATC-3', and an antisense primer: 5'CGAGCTCTTCAAAAAGTTGCATGGTGCTGG-3'. Amplification was performed for 30 cycles using the Platinum Pfx DNA Polymerase. The PCR hot-start procedure was as follows: 95°C for 6 min, 94°C for 40 s, 68°C for 3 min, plus 1 min after each 10 cycles, and 68°C for 10 min. The HindIII/EcoRI-digested PCR products were ligated into HindIII/EcoRI-digested pcDNA3.1 (+) vector using T4 DNA ligase. The recombinant plasmids were then transformed into Escherichia coli (E. coli) DH5α and confirmed by restriction endonuclease digestion and DNA sequence analysis. The sequences were aligned using the Gene Runner version 3.05 (Hastings Softerware Inc., Hastings, NY, USA).
HepG2 cells were cultured in DMEM, supplemented with 10% fetal calf serum, penicillin (100 U/mL) and streptomycin (100 g/mL) at 37°C in a humidified incubator with 5% CO2. HepG2 cells in the exponential phase of growth were strictly counted and seeded onto 24-well culture plates with 1.0 × 105 cells/well. After 24 h, cells at 80%-90% confluence were transfected with the recombinant plasmids using Lipofection 2000 reagent, following the manufacturer’s guidelines. The transfected cells and supernatants were then harvested after 48 or 72 h. Vector pcDNA3.1 (+) was used as a mock transfection control.
At 48 or 72 h post-transfection, the culture supernatant was collected, centrifuged at 3000 r/min for 5 min to remove cellular debris, and transferred to a clean tube for further analysis. The expression levels of HBsAg and HBeAg were separately assayed using an enzyme immunoassay kit. According to the instructions, a ratio of sample/negative (S/N) ≥ 2.1 was considered as a positive response to HBsAg or HBeAg antigen.
Real-time fluorimetry PCR using TaqMan probe was performed to quantify HBV DNA at 48 or 72 h post-transfection. HBV DNA was extracted from the intracellular core particles using Axygen DNA Mini kit, and then examined by Quantitative HBV PCR Fluorogence Diagnostic kit. According to the instructions, an HBV DNA level ≥ 5.0 × 102 copies/mL was considered as a positive response.
All experiments were performed at least three times. All data were indicated as mean ± SD. Data analysis was performed by SPSS 10.0 software (Spss Inc., Chicago, IL, USA).
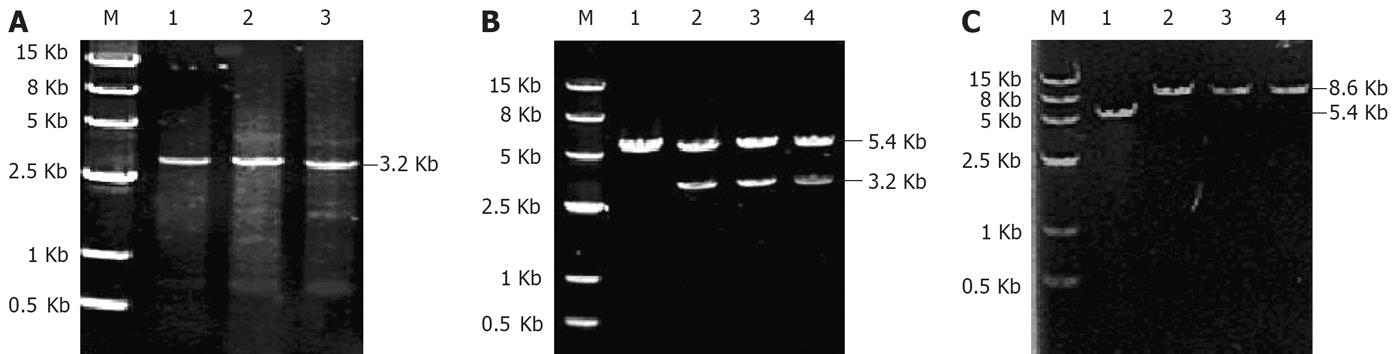
As shown in Figure 1A, the PCR products had the expectant molecular weight (3.2 kb). The target genes were cloned to the expression vector pcDNA3.1 (+), and transformed into E. coli DH5α, which generated the reconstructed plasmids pcDNA3.1 (+)/HBV/C-YMDD, YIDD or YVDD. After amplification by E. coli, the recombinant plasmids were extracted from the positive clones, and then characterized by digestion with restriction enzymes HindIII/EcoRI (Figure 1B, lanes 2-4) and EcoRI (Figure 1C, lanes 2-4). Vector pcDNA3.1 (+), used as a negative control, was also digested with EcoRI,which yielded a product of approximate 5.4 kb in size (Figure 1B and C, lane 1). The digested products of the recombinant plasmids were visualized on 7 g/L agarose gel (Figure 1B), which demonstrated that recombinant plasmids were digested to 5.4 and 3.2 kb DNA fragments, which corresponded to the lined vector pcDNA3.1 (+) (5.4 kb) and the target gene HBV full-length genome (3.2 kb), respectively. As shown in Figure 1C, the fragment digested from the recombinant plasmids by EcoRI was approximate 8.6 kb in size, as expected.
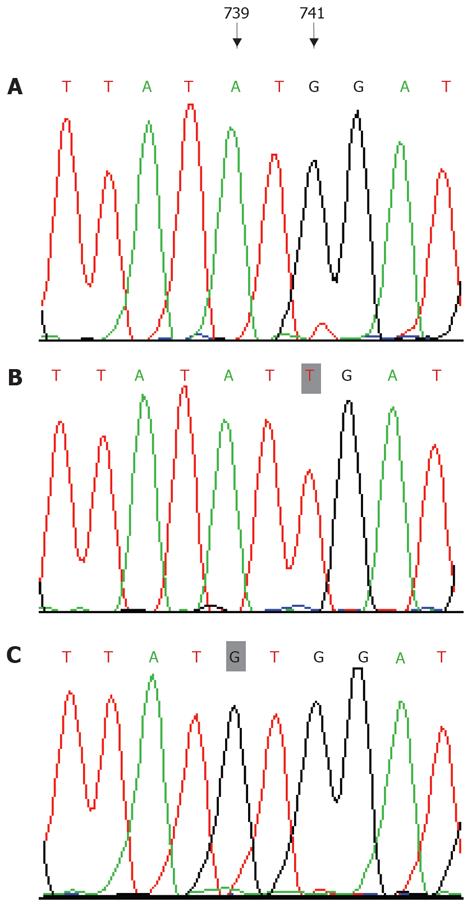
DNA sequence analysis of positive clones confir-med the result. The inserted HBV full-length genome had the correct reading frame and length. Compared with the sequence of the recombinant plasmids that contained wild-type strain (Figure 2A), it was clearly shown that, in the HBV YIDD mutant (Figure 2B), the 741th base G mutated to T, and in the HBV YVDD mutant (Figure 2C), the 739th base A mutated to G. These mutations resulted in replacement of the methionine residue (amino acid 204) by isoleucine (rtM204I), or valine (rtM204V), respectively.
At 48 or 72 h post-transfection, culture supernatants were collected. The expression levels of HBV HBsAg and HBeAg were then detected by ELISA. According to the instructions, an S/N ratio ≥ 2.1 was considered as positive HBeAg response. As shown in Table 1, our results indicated that each of the recombinant plasmids could express the antigens, HBsAg and HBeAg in HepG2 cells. The blank control group had a negative HBsAg and HBeAg response.
| Clone | HBsAg (Sample/Negative) | HBeAg (Sample/Negative) | ||
| 48 h | 72 h | 48 h | 72 h | |
| pcDNA3.1 (+) | 0.33 ± 0.028 | 0.37 ± 0.094 | 0.39 ± 0.046 | 0.38 ± 0.050 |
| pcDNA3.1 (+)-HBV/C YMDD | 3.14 ± 0.069 | 3.47 ± 0.413 | 8.72 ± 0.059 | 8.77 ± 0.256 |
| pcDNA3.1 (+)-HBV/C YIDD | 6.77 ± 0.099 | 8.26 ± 0.334 | 2.06 ± 0.318 | 2.18 ± 0.028 |
| pcDNA3.1 (+)-HBV/C YVDD | 10.30 ± 0.065 | 10.37 ± 0.205 | 5.03 ± 0.132 | 5.30 ± 0.117 |
At 48 or 72 h post-transfection, HepG2 cells were harvested and real-time fluorimetry PCR was then performed. As shown in Table 2, the three transfection groups could be considered as positive (all ≥ 5.0 × 102 copies/mL), which indicated that HBV DNA was expressed efficiently. The blank control group was negative.
Lamivudine, a potent inhibitor of HBV replication has been the main therapeutic option for treatment of chronic hepatitis B. It functions by interfering with HBV reverse transcriptase activity, and leads to a marked decrease in serum HBV DNA levels, a significant increase in the rate of HBeAg seroconversion, as well as improvement in serum alanine aminotransferase (ALT) levels[15] and liver histopathological parameters[16]. Several data have revealed that lamivudine can efficiently promote the treatment of hepatitis B in the short term. However, the long-term effectiveness of lamivudine is hampered by the development of viral resistance[1517]. Lamivudine resistance is associated with mutations in the highly conserved YMDD motif of the reverse transcriptase, in which, methionine 204 is replaced by either isoleucine (rtM204I, YIDD variant) or valine (rtM204V, YVDD variant).
It has been reported that the rate of HBV YMDD mutation increases with the duration of lamivudine therapy, with an increase from 15% in one year to 38% and 53% after two and three years of treatment, respectively[18]. Our previous research has also indicated that in northern China, the YMDD mutation rate is approximate 56.3% after four years of lamivudine treatment[8]. YMDD mutations not only result in a reduction in the susceptibility to lamivudine, but also cause virological and biochemical breakthrough, which are represented as rebound of HBV DNA and ALT levels[1920]. Moreover, acute exacerbation of hepatitis and hepatic failure may occur after the emergence of YMDD mutants. Therefore, the antiviral treatment of YMDD mutants has become a crucial issue in the clinic.
HBV genotypes have distinct geographical distributions and are potential factors that affect virus replication, virus variation, clinical course, and therapy of HBV infection. In northern China, genotype C is predominant, and accounts for 77%-88% of cases of chronic hepatitis B[68]. Sugiyama reported that the replication capacity of HBV in transfected Huh7 cells varied among genotype A and B, as well as C and D, with genotype C having the highest replication capacity[21]. HBV genotype C is associated with more severe histological liver damage and low-grade responses to interferon therapy[22]. Moreover, patients with genotype C show poor responses to embolization therapy and may die from hepatic failure because of rapid hepatocellular carcinoma (HCC) progression[23]. Another study has reported that HBV genotype C has more rapid selection of lamivudine resistance than genotype B[17].Therefore, further studies of HBV YMDD mutants with genotype C are of great significance.
To date, many in vitro studies on lamivudine resistance have been reported[24–27]. In most of these, recombinant plasmids containing HBV full-length or fragment genome were constructed first, and then expressed in liver-derived cell lines. For example, Gunther et al have reported an original and efficient method of amplifying full-length HBV genomes by PCR[24]. Chen et al have described a method of constructing baculovirus recombinants that contain multiple HBV lamivudine-resistant mutations, introduced by successive rounds of site-directed mutagenesis in laboratory strains[25]. However, in all these studies, either one type of HBV YMDD mutant or wild-type strains was included in the plasmids without specification of HBV genotype. Therefore, to date, serial plasmids that contain a specific HBV genotype, such as genotype C, and lamivudine-resistant sequences, which allow systematic studies on the combined effects of HBV genotype together with lamivudine-resistant mutations, have not been reported.
In this study, we successfully constructed a series of eukaryotic expression plasmids that contained genotype C HBV strain with either wild-type, YVDD or YIDD mutation, namely the plasmids pcDNA3.1 (+)-HBV/C-YMDD, pcDNA3.1 (+)-HBV/C-YVDD and pcDNA3.1 s(+)-HBV/C-YIDD, respectively. In order to achieve high-level expression in vitro, the Kozak sequence, ACCATGGCC-which has been found to contribute to the fidelity and efficiency of initiation and expression[28]-was coupled to the 5' end of the sense primer. Moreover, to further assure high fidelity, the PCR analyses were performed following a hot-start protocol and using high-fidelity enzymes. After transfecting the constructed plasmids into HepG2 cells, we analyzed the expression levels of HBsAg and HBeAg by ELISA, and the replication level of HBV DNA by real-time PCR. It was found that both HBV DNA and the antigens were expressed in the transfected cells, but not in the negative control cells transfected with pcDNA3.1 (+). As shown in Table 1, all the recombinant plasmids could express HBsAg in HepG2 cells. At 48 and 72 h, the expression levels of HBeAg were 8.723 ± 0.0585 and 8.77 ± 0.256, respectively, in YMDD strains and 5.03 ± 0.132 and 5.3 ± 0.117 in YVDD mutants. However, HBeAg expression levels in YIDD mutants were only 2.06 ± 0.318 and 2.18 ± 0.028 at 48 and 72 h, respectively. This difference was probably caused by the emergence of BCP mutations (A1762T/G1764A) in YIDD mutants, while this mutation was not present in YVDD mutants. Our observation is consistent with previous reports that BCP mutation can result in a decrease in HBeAg levels[29]. In addition, HBV DNA expression levels of each of the recombination plasmids were ≥ 108 copies/mL in HepG2 cells (Table 2), which indicates that the three recombinant plasmids can be expressed efficiently. Successful construction of the three eukaryotic plasmids pcDNA3.1 (+)-HBV/C-YMDD, pcDNA3.1 (+)-HBV/C-YVDD and pcDNA3.1 (+)-HBV/C-YIDD, provides an experimental basis for the establishment of stable expression system of HBV genotype C lamivudine-resistant mutants. The results may contribute to future in vitro antiviral studies of HBV genotype C lamivudine-resistant mutants.
HBV infection remains a major public health problem worldwide. Lamivudine is currently one of the most effective anti-hepatitis B virus (HBV) drugs in use clinically. However, the long-term use of lamivudine leads to the emergence of lamivudine-resistant mutants (YMDD mutants). It was reported that the rate of YMDD mutations was up to 70% after three years of treatment. The development of YMDD mutants has hampered anti-HBV therapy.
In vivo and in vitro studies on the HBV drug-resistance mechanism have been of great interest. In vivo studies have mainly focused on the rate, types and detection method of YMDD mutation. However, there is still little known about the effects of YMDD mutations in vitro.
Appropriate and effective eukaryotic expression plasmids that are able to efficiently express HBV DNA and antigens are necessary for further in vitro investigations. However, to date, serial plasmids that contain a specific HBV genotype, such as genotype C, and a certain lamivudine-resistance mutation, which allow systematic studies of the combined effects of HBV genotype, together with lamivudine-resistance mutations, have not been reported. In this study, authors successfully constructed eukaryotic expression plasmids pcDNA3.1 (+)-HBV/C-YMDD, pcDNA3.1 (+)-HBV/C-YVDD and pcDNA3.1 (+)-HBV/C-YIDD, which contained genotype C HBV strain with either wild-type, YVDD or YIDD mutations, respectively, and had the ability to express HBV DNA and antigens in vitro with a high capacity.
The successful construction of three eukaryotic plasmids, pcDNA3.1 (+)-HBV/C-YMDD, pcDNA3.1 (+)-HBV/C-YVDD and pcDNA3.1 (+)-HBV/C-YIDD, provides an experimental basis for the establishment of a stable expression system of HBV genotype C lamivudine-resistant mutants. The results may contribute to further in vitro antiviral studies of HBV genotype C lamivudine-resistant mutants. This could include establishing a stable expression system for HBV genotype C lamivudine-resistant mutants for studying the mechanism of HBV lamivudine resistance.
HBV genotype C is predominant in China, and is associated with more severe histological liver damage, lower response to anti-HBV treatment, and more rapid development of lamivudine resistance.
The paper describes a technique for constructing eukaryotic expression plasmids of HBV genotype C with lamivudine-resistant mutants. This is an interesting topic and the manuscript is well written.
| 1. | Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329-338. |
| 2. | Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67-74. |
| 3. | Kao JH, Chen PJ, Lai MY, Chen DS. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J Clin Microbiol. 2002;40:1207-1209. |
| 4. | Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-2073. |
| 5. | Jazayeri M, Basuni AA, Sran N, Gish R, Cooksley G, Locarnini S, Carman WF. HBV core sequence: definition of genotype-specific variability and correlation with geographical origin. J Viral Hepat. 2004;11:488-501. |
| 6. | Gu HX, Xu ZL, Liu JY, Zhong ZH, Wang HQ, Zhang SY, Li D, Zhang HH, Abe K. Epidemiology of HBV genotypes by nested PCR with multi-paired primers. Shijie Huaren Xiaohua Zazhi. 2004;12:1073–1076. |
| 7. | Ding X, Mizokami M, Yao G, Xu B, Orito E, Ueda R, Nakanishi M. Hepatitis B virus genotype distribution among chronic hepatitis B virus carriers in Shanghai, China. Intervirology. 2001;44:43-47. |
| 8. | Li D, Gu HX, Zhang SY, Zhong ZH, Zhuang M, Hattori T. YMDD mutations and genotypes of hepatitis B virus in northern China. Jpn J Infect Dis. 2006;59:42-45. |
| 9. | Ding X, Gu H, Zhong ZH, Zilong X, Tran HT, Iwaki Y, Li TC, Sata T, Abe K. Molecular epidemiology of hepatitis viruses and genotypic distribution of hepatitis B and C viruses in Harbin, China. Jpn J Infect Dis. 2003;56:19-22. |
| 10. | Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL, Brown N, Condreay LD. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670-1677. |
| 11. | Stuyver L, Van Geyt C, De Gendt S, Van Reybroeck G, Zoulim F, Leroux-Roels G, Rossau R. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J Clin Microbiol. 2000;38:702-707. |
| 12. | Sun J, Wang Z, Ma S, Zeng G, Zhou Z, Luo K, Hou J. Clinical and virological characteristics of lamivudine resistance in chronic hepatitis B patients: a single center experience. J Med Virol. 2005;75:391-398. |
| 13. | Pallier C, Castera L, Soulier A, Hezode C, Nordmann P, Dhumeaux D, Pawlotsky JM. Dynamics of hepatitis B virus resistance to lamivudine. J Virol. 2006;80:643-653. |
| 14. | Suzuki Y, Yotsuyanagi H, Okuse C, Nagase Y, Takahashi H, Moriya K, Suzuki M, Koike K, Iino S, Itoh F. Fatal liver failure caused by reactivation of lamivudine-resistant hepatitis B virus: a case report. World J Gastroenterol. 2007;13:964-969. |
| 15. | Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Wu PC, Dent JC, Barber J. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61-68. |
| 16. | Dienstag JL, Goldin RD, Heathcote EJ, Hann HW, Woessner M, Stephenson SL, Gardner S, Gray DF, Schiff ER. Histological outcome during long-term lamivudine therapy. Gastroenterology. 2003;124:105-117. |
| 17. | Pan XP, Li LJ, Du WB, Li MW, Cao HC, Sheng JF. Differences of YMDD mutational patterns, precore/core promoter mutations, serum HBV DNA levels in lamivudine-resistant hepatitis B genotypes B and C. J Viral Hepat. 2007;14:767-774. |
| 18. | Leung NW, Lai CL, Chang TT, Guan R, Lee CM, Ng KY, Lim SG, Wu PC, Dent JC, Edmundson S. Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: results after 3 years of therapy. Hepatology. 2001;33:1527-1532. |
| 19. | Yuen MF, Sablon E, Hui CK, Yuan HJ, Decraemer H, Lai CL. Factors associated with hepatitis B virus DNA breakthrough in patients receiving prolonged lamivudine therapy. Hepatology. 2001;34:785-791. |
| 20. | Wang JH, Lu SN, Lee CM, Lee JF, Chou YP. Fatal hepatic failure after emergence of the hepatitis B virus mutant during lamivudine therapy in a patient with liver cirrhosis. Scand J Gastroenterol. 2002;37:366-369. |
| 21. | Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N. Influence of hepatitis B virus genotypes on the intra-and extracellular expression of viral DNA and antigens. Hepatology. 2006;44:915-924. |
| 22. | Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg (+) chronic hepatitis than genotype C. Hepatology. 2002;36:1425-1430. |
| 23. | Tsubota A, Arase Y, Ren F, Tanaka H, Ikeda K, Kumada H. Genotype may correlate with liver carcinogenesis and tumor characteristics in cirrhotic patients infected with hepatitis B virus subtype adw. J Med Virol. 2001;65:257-265. |
| 24. | Gunther S, Li BC, Miska S, Kruger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437-5444. |
| 25. | Chen RY, Edwards R, Shaw T, Colledge D, Delaney WE 4th, Isom H, Bowden S, Desmond P, Locarnini SA. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology. 2003;37:27-35. |
| 26. | Sun D, Nassal M. Stable HepG2- and Huh7-based human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus. J Hepatol. 2006;45:636-645. |
| 27. | Brunelle MN, Jacquard AC, Pichoud C, Durantel D, Carrouee-Durantel S, Villeneuve JP, Trepo C, Zoulim F. Susceptibility to antivirals of a human HBV strain with mutations conferring resistance to both lamivudine and adefovir. Hepatology. 2005;41:1391-1398. |
| 28. | Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887-903. |