Published online Jun 14, 2008. doi: 10.3748/wjg.14.3526
Revised: May 6, 2008
Accepted: May 13, 2008
Published online: June 14, 2008
AIM: To establish the role of FHIT in the pathogenesis hepatocellular carcinoma (HCC).
METHODS: We examined genomic alterations, as well as, mRNA and protein expression patterns from the FHIT gene, in 48 surgically resected hepatocellular carcinoma (HCC) tissues. Additionally, p53 mutations were analyzed.
RESULTS: Aberrant FHIT transcripts were detected in 11 of 48 surrounding non-tumor liver tissues and 27 of 48 HCC samples (22.9% vs 56.3%, P = 0.002). No point mutations were identified within the open reading frame region of FHIT. Loss of heterozygosity (LOH) of the FHIT locus was detected in 4 of 42 informative cases for D3S1300, and 3 of 29 informative cases for D3S1313. Reduced expression of FHIT protein (Fhit) was observed in 8 (16.7%) of 48 HCC samples, with complete loss of Fhit in only 1 case. There were no associations with abnormal transcripts, LOH, and Fhit expression. p53 mutations were identified in 9 of the 48 HCC cases. However, none of the cases displayed a G to T transversion at p53 codon 249.
CONCLUSION: Aberrant FHIT transcripts were more common in HCC tissues as compared to non-cancerous liver tissues. However, Fhit expression was lost or reduced in a minor fraction of HCC tissues, while it was strongly expressed in non-cancerous liver tissues. Therefore, our study suggests that FHIT plays a role in relatively few HCC cases in South Korea.
- Citation: Nam CW, Shin JW, Park NH. Fragile histidine triad gene alterations are not essential for hepatocellular carcinoma development in South Korea. World J Gastroenterol 2008; 14(22): 3526-3533
- URL: https://www.wjgnet.com/1007-9327/full/v14/i22/3526.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.3526
Hepatocellular carcinoma (HCC) is currently the fifth most common cancer worldwide and the fourth leading cause of cancer-related deaths. The number of new cases is estimated as more than 500 000 per year, accounting for 4% of all newly diagnosed cancers[1]. More than 80% of HCC cases occur in developing countries, especially in South-East Asia and sub-Saharan Africa, but the incidence is increasing in economically developed regions, including Japan, Western Europe, and the United States[12]. The overall prognosis of HCC is poor, because many patients at presentation are already in an advanced and unresectable state, and will have a median survival time of less than 6 mo[3]. High mortality may be partially attributable to the fact that the nanocapsular part of the liver lacks sensory fibers, leading to symptoms presentation only in the advanced HCC[3]. Therefore, only a small proportion of patients are eligible for liver resection, which results in a 5-year survival rate of about 40%[4]. However, even following surgical resection, recurrence rates can be as high as 50% at 2 years[4].
Hepatocarcinogenesis is a multistep process involving different genetic alterations that ultimately lead to the malignant transformation of hepatocytes. Numerous genetic and epigenetic alterations contribute to the activation of carcinogenic pathways in HCC[4]. Whereas most studies on the mechanisms of tumori-genesis have focused on genetic changes, various epigenetic changes have been increasingly identified in HCC[4]. The short arm of human chromosome 3 is one of the most common sites of chromosomal abnormality in malignant diseases[5–8]. One candidate is the fragile histidine triad (FHIT) gene, located on 3p14.2, spanning the FRA3B common fragile site[910]. The FHIT gene is composed of 10 exons encompassing 1.8 Mb genome regions, of which only exons 5 to 9 code for protein. It encodes a small mRNA of 1.1 kb, and a small protein of 16.8 kDa[9]. Studies on the functional aspects of FHIT have been reported that this gene is a bona fide tumor suppressor gene[11]. The FHIT gene and its protein have may be involved in the regulation of cell proliferative and apoptotic processes[11]. Down-regulation of FHIT inhibits apoptosis and FHIT protein (Fhit) interacts with a number of key proteins involved in cancer progression, including p53[1112]. Coexpression of FHIT and p53 synergistically inhibits the proliferation of various tumor cell lines. These synergistic effects may occur because of stabilization of p53 related to Fhit-mediated downregulation of MDM2[12]. Altered transcripts and allelic loss of the FHIT gene are frequently identified in premalignant and malignant lesions of various tumors[13–18]. Moreover, loss or reduction of Fhit expression has been found in most tumors including HCC[10–1419–22].
To establish the role of FHIT in the pathogenesis of HCC, we examined genomic alterations, as well as, mRNA and protein expression in surgically resected HCCs and their associated non-cancerous surrounding liver tissues. We also investigated the possible associations between FHIT abnormalities and p53 mutations.
HCC samples and their corresponding non-cancerous liver tissues were obtained from 48 patients who had undergone surgical resection at Ulsan University Hospital in South Korea. Written informed consent was obtained from all patients participating in this study, and the study was approved by the Institutional Review Boards at Ulsan University Hospital. All cancer samples were obtained from within the tumor. Matching surrounding non-cancerous liver tissues were obtained as far as possible from the tumors. All specimens were frozen immediately after surgical resection, and stored at -70°C. HCC diagnosis was based on histologic confirmation or elevated serum AFP (> 400 ng/mL) with radiologic findings, or at least two coincident radiologic findings (contrast-enhanced dynamic computer tomography (CT), contrast-enhanced dynamic magnetic resonance imaging (MRI), and Doppler ultrasonography compatible with HCC.
Postoperative follow-up included a dynamic CT/MRI study every 3 mo, and laboratory testing of the serum AFP level every 1 to 3 mo at our outpatient clinic. In the case of suspected HCC recurrence, further examinations, including angiography and lipiodol CT, were performed. If necessary, ultrasound-guided biopsy was conducted to confirm the diagnosis. Bone scintigraphy or chest CT was performed when clinically indicated. The follow-up period was defined as the interval from the date of surgical resection of HCC until the date diagnosis of the recurrence, or the date of death, or the end of follow-up.
Total RNA and DNA were isolated from the HCCs and corresponding non-cancerous tissues, using the TRIZOL reagent (Gibco BRL Life Technologies, Gaithersburg, MD, USA). cDNA was synthesized by reverse transcriptase from 1 &mgr;g of total RNA. One ul of the RT reaction was used to amplify the FHIT cDNA as described by Ohta et al[9]. To amplify FHIT exons 3-10, a nested PCR was carried out in 10-&mgr;L final volume with 30 ng of primers (5U2-3D2 and 5U1-3D1, according to Ohta et al[9]. The first round of PCR amplification was performed in 10 uL of reaction mixture comprising 1 &mgr;mol/L primers 5U2-3D2 and 5U1-3D1, 200 &mgr;mol/L each dNTP, 1 × PCR buffer, 2.5 U of ampliTaq, and 0.5 &mgr;L of the synthesized cDNA mixture under the following reaction conditions: denaturation for 10 min at 95°C, 34 cycles of 30 s at 95°C, 30 s at 56°C, and 45 s at 72°C, and a final extension step for 10 min at 72°C. After 10-fold dilution of the amplified product 10-fold in DW, 2 &mgr;L of aliquots were subjected to a second round of PCR amplification using nested primers, 5U1 and 3D1, under the above conditions. Each nested RT-PCR assay was repeated at least twice with the original extracted RNA for confirmation. All reactions were performed at least twice and the integrity of the RNA samples verified. To confirm the presence of mRNA and rule out nonspecific amplification, 25 cycles of ®-actin cDNA were preformed after RT, both with and without the RT enzyme. PCR products were directly sequenced using primer 5U1 and 3U1 with an automated Applied Biosystem Model 3730 DNA sequencer (Perkin-Elmer, Foster City, CA, USA). For sequence analysis of abnormal FHIT transcripts, the PCR products were resolved in 1.5% ethidium bromide agarose gel. Bands were excised from gels, and PCR products were purified using the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA, USA). PCR products excised from gels were directly sequenced using the sequencing primers described by Zekri et al[18].
To investigate the allelic loss of FHIT gene, a PCR-based approach was performed utilizing primers that amplify two polymorphic microsatellite markers internal to and flanking the FHIT gene. The microsatellite marker D3S1300 is located in intron 5 of the FHIT gene, whereas marker D3S1313 is slightly telomeric to FHIT. The 10 &mgr;L PCR mixture contained 1 × Gold Buffer, 0.8 &mgr;L 25 mmol/L MgCl2, 0.8 &mgr;L 10 mmol/L dNTP, 1 &mgr;L 5 &mgr;mol/L concentrations of each primer pair, 0.5 &mgr;L 0.25 unit Amplitaq Gold, and 2 &mgr;L of the 10 ng extracted DNA. 1 &mgr;L of diluted PCR product was mixed with 9 &mgr;L of loading buffer (formamide: Ro × 500 size standard, 1:39). The mixture was denatured at 95°C for 5 min, chilled on ice, and loaded on an ABI PRISM 3100 automatic sequencer. The data were analyzed using the GeneScan and Genotyper software (Perkin-Elmer/Applied Biosystems). Constitutional homozygosity was regarded as uninformative. For informative cases, allelic loss was scored as positive if the signal of one allele was lost or reduced to at least 50% in tumor DNA, compared with the corresponding normal allele. All samples were run twice to confirm the presence or absence of allelic imbalance.
Protein concentration was measured using the BCA protein assay (Pierce, Rockford, IL). Ten micrograms of lysate protein was separated by SDS-PAGE using a 12% polyacrylamide gel and electroblotted onto a Hybond ECL (Amersham Pharmacia Biotech). After blockage of nonspecific binding sites for 1 h with 5% nonfat milk in TPBS (PBS and 0.1% Tween 20), the membrane was incubated at room temperature for 2 h with rabbit anti-FHIT antibody (Zymed Laboratories, South San Francisco, CA, USA) at a dilution of 1:500. The membrane was washed three times with TPBS, incubated further with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (New England Biolabs, Beverly, MA, USA) at room temperature, and then washed three times with TPBS. Membranes were developed with the ECL chemiluminescence system (Amersham Pharmacia Biotech). The intensity of Fhit expression in HCC tumors was compared with that in non-cancerous surrounding liver tissues.
The primers used were oligonucleotides complementary to the sequence flanking the exon/intron junctions of exons 5-9. The sequence of the primers is as follows: exon 5, 5'-CTGACTTTCAACTCTG-3' (forward) and 5'-AGCCCTGTCGTCTCT-3' (reverse); exon 6, 5'-CTC TGATTCCTCACTG-3' (forward) and 5'-ACCCCAGTTGCAAACC-3' (reverse); exon 7, 5'-TGCTTGCCACAGGTCT-3' (forward) and 5'-ACAGCAGGCCAGTGT-3' (reverse); exon 8, 5'-AGGACCTGATTTCCTTAC-3' (forward) and 5'-TCTGAGGCATAACTGC-3' (reverse); and exon 9, 5'-TATGCCTCAGATTCACT-3' (forward) and 5'-ACTTGATAAGAGGTCC-3' (reverse). The same primers sets were used for DNA sequencing. PCR was carried out in a 20-&mgr;L reaction contained; 50 ng DNA, 10 × PCR buffer, 200 &mgr;mol/L each of dNTP, 5 pmol of each primer and 1 U of Taq DNA polymerase. PCR conditions were as follows: 95°C (10 min) for 1 cycle, 95°C (40 s), 63°C (40 s; for exons 4, 5, and 7-9) or 67°C (40 s; for exon 6), 72°C (40 s) for 40 cycles, and a final extension step of 72°C (10 min). PCR products were separated by electro-phoresis and visualized with 1.5% ethidium bromide. Samples without the DNA template were included in all assays as negative controls. PCR products were purified using the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA, USA), and they were sequenced by the dideoxychain termination method with the Big Dye Terminator cycle sequencing kit (Perkin-Elmer Corporation, Foster City, CA, USA). Cycle sequencing was performed for 25 cycles of denaturation (96°C, 30 s), annealing (50°C, 15 s), and extension (60°C, 4 min), according to the Big Dye Terminator protocol. After spin column purification with Centri-Sep columns (Perkin-Elmer Corporation), samples were analyzed with the automated Applied Biosystem Model 3730 DNA sequencer (Perkin-Elmer, Foster City, CA, USA).
All continuous variables were compared using the Mann-Whitney test, or the one-way analysis of variance (ANOVA). Categorical variables were compared using the Fisher’s exact test. All data were analyzed using the statistical package SPSS (version 14.0: SPSS Inc., Chicago, IL, USA). In all cases, a 2-tailed P value of less than 0.05 was considered statistically significant.
The baseline characteristics of the 48 patients were shown in Table 1. The patients included 35 men and 13 women with a mean age of 50 years (range 28-71 years). HBV was the most common etiology, accounting for 38 (79%) of the 48 cases. Antibody to HCV was identified in three cases. Seven patients were negative for HBV and HCV. Alpha-fetoprotein (AFP) level ranged from 1.0 to 240 000 ng/mL with a median level of 202 ng/mL. Twelve (25%) of the 48 patients had normal AFP levels (< 11 ng/mL), whereas 20 (41.7%) patients had AFP levels of > 400 ng/mL. Preoperative diagnosis of HCC was based on histologic confirmation in 28 cases, or elevated serum AFP (> 400 ng/mL) with radiologic findings, or at least two coincident radiologic findings compatible with HCC in the remaining 20 cases. In 22 of the 48 patients, cirrhosis was observed in the surrounding non-cancerous liver tissue, and chronic hepatitis was identified in parenchyma surrounding the tumors in the remaining 26 cases. According to the Child-Pugh classification at the time of operation, 19 patients were class A (CPT 5-6), 3 were class B. Based on UICC TNM staging[23], 5 patients were stage I, 41 stage II, and 2 stage III. Small HCC was defined as a single tumor < 5 cm, or 2-3 tumors < 3 cm without invasion of major veins larger than sub-segmental branches. These criteria are similar to those applied to define early stage HCC in the BCLC (Barcelona Clinic Liver Cancer) staging scheme[24]. Tumors exceeding these limits were regarded as advanced HCC. At initial diagnosis, 26 patients had small HCC, while 22 patients were advanced HCC. The mean tumor size was 5.8 ± 3.9 cm. Among the 46 patients with solitary tumors, a diameter of 3 cm or less was observed in 11 (23.9 %), between 3 cm and 5 cm in 15 (32.6%), and greater than 5 cm in 20 (43.5%). There were no tumors with positive lymph nodes or macroscopic vascular invasion by HCC. Of these HCCs, 8 were at grade I, 13 at grade II, 20 at grade III and 5 at grade IV according to the Edmondson and Steiner’s grade[25]. Microscopically, vascular and tumor capsular invasions were detected in 13 (27%) and 5 (10%) patients, respectively.
| Characteristics | |
| Mean age (yr) | 50 (20-71) |
| Sex (M:F) | 35:13 |
| Etiology of liver disease | |
| HBV | 38 |
| HCV | 3 |
| Non-HBV, Non-HCV | 7 |
| Tumor size | |
| ≤ 3 cm | 11 |
| 3-5 cm | 15 |
| > 5 cm | 22 |
| Mean ( range) (cm) | 5.8 ± 3.9 |
| UICC TNM stage | |
| I | 5 |
| II | 41 |
| III | 2 |
| Edmonson-Steiner’s grade | |
| I | 8 |
| II | 13 |
| III | 20 |
| IV | 5 |
| Underlying liver disease | |
| Cirrhosis | 22 |
| Chronic hepatitis | 26 |
| AFP (ng/mL) | |
| < 11 | 12 |
| 11-400 | 16 |
| ≥ 400 | 20 |
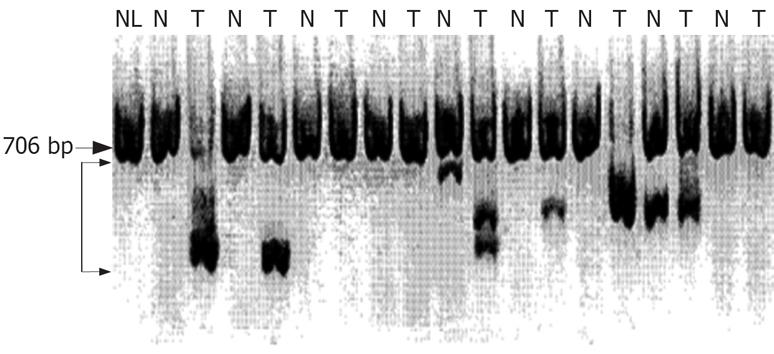
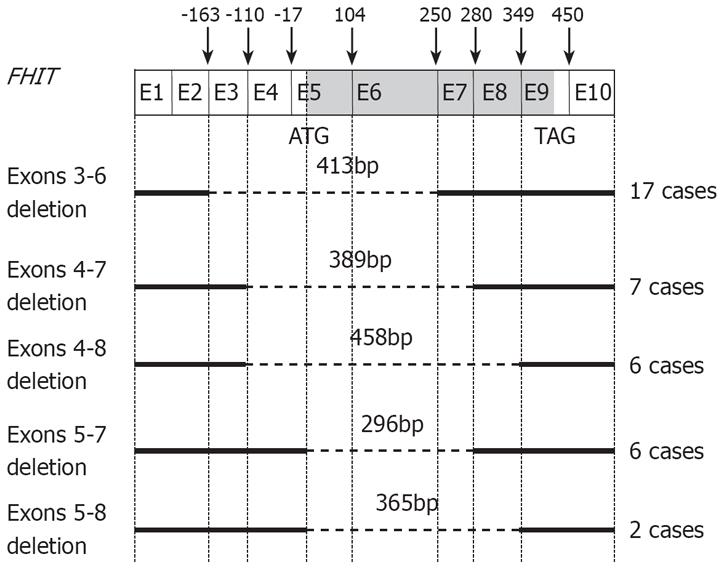
Representative results of nested RT-PCR analysis are shown in Figure 1. Abnormal-sized bands representing aberrant RT-PCR products were detected in 11 (22.9%) of 48 surrounding non-tumorous liver tissues and 27 (56.3%) of the 48 HCC samples. Thus, the number of tumors with aberrant transcripts was greater than that of non-tumor tissues with aberrant transcripts (P = 0.002). Such abnormal-sized transcripts were seen at five different positions at 296, 365, 389, 413, and 458 bp (Figure 1). Interestingly, 13 cases displayed two or three aberrant transcripts. Sequence analysis of the aberrant transcripts revealed deletions of exons 3-6 (nt-164 to 249) in 17 cases, exons 4-7 (nt-110 to 279) in 7 cases, exons 4-8 (nt-110 to 348) in 6 cases, exons 5-7 (nt-17 to 279) in 6 cases, and exons 5-8 (nt-17 to 348) in 2 cases (Figure 2). Among 11 surrounding non-tumorous liver tissue samples with aberrant transcripts, 5 were chronic hepatitis, whereas in the remaining 6 cases, the parenchyma surrounding the tumor was cirrhosis. In total, 8 patients displayed aberrant transcripts in both tumor and surrounding non-tumor tissues, 5 of whom displayed the same abnormal patterns in the paired tumor and non-tumor samples; however, in the remaining 3 cases, the pattern was different between the paired samples. Whereas 19 cases had aberrant transcripts only in their tumor tissues, we also found 3 cases showing an aberrant transcript in their non-tumor tissues only. However, no point mutations of the FHIT gene were found in the region of the open reading frame. All normal-sized transcripts of 706 bp exhibited wild-type sequences. All of 21 tumor tissues without aberrant transcripts were presented normal sized transcripts. Normal-sized transcripts were also observed in 22 of the 27 HCCs with aberrant FHIT transcripts, but were barely present or completely absent in the remaining 5 cases. However, normal-sized bands were observed in all surrounding non-tumor tissues samples, regardless of the presence of aberrant transcripts. The presence of FHIT aberrations was not associated with clinical parameters, including age, sex, Child-Pugh classification, histological grade of tumor, presence of tumor capsule, AFP, tumor size, microscopic invasion, tumor recurrence, or survival (P > 0.05).
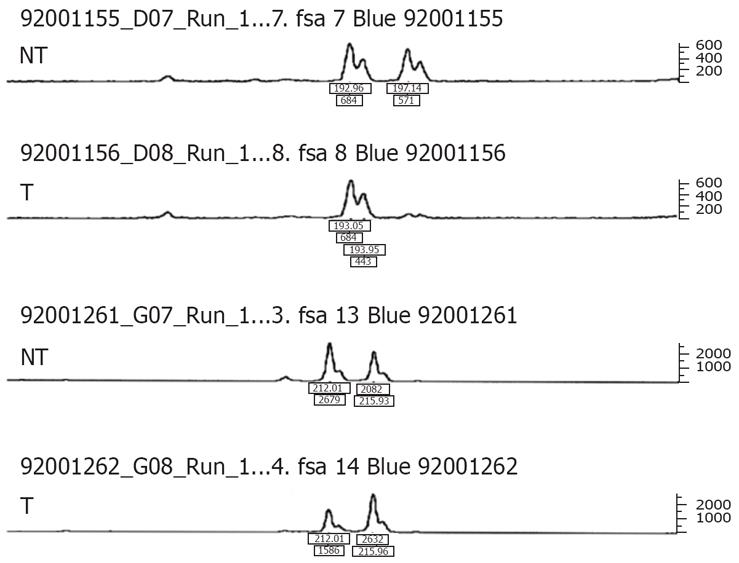
Among the 48 HCCs, 7 displayed LOH of D3S1300, D3S1313, or both markers. Specially, 4 of the 42 informative cases at D3S1300, and 3 out of the 29 cases informative at D3S1313, exhibited LOH. LOH involving both allelic markers was not observed. Examples are shown in Figure 3. In total, 6 of 30 cases with aberrant transcripts, and only 1 of 18 cases without aberrant transcripts showed LOH. Thus, the incidence of LOH appeared higher in cases with aberrant transcripts than cases without aberrant transcripts, although the data were not statistically significant (P = 0.23). Moreover, we observed a trend towards FHIT aberration in tumors with LOH, although again the difference was not statistically significant (P = 0.23). LOH of the FHIT gene was not associated with the clinical parameters examined.
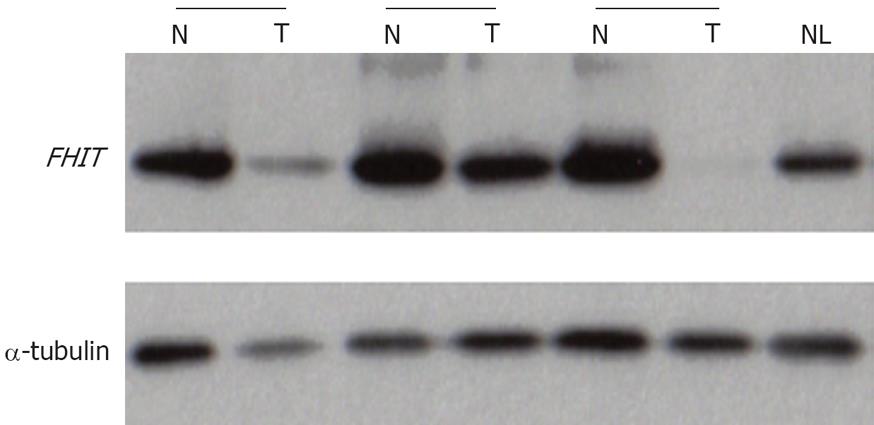
Representative results of Western blot analysis are shown in Figure 4. Eight (16.7%) of 48 HCCs exhibited reduced or no Fhit expression, compared with normal liver tissues. Complete loss of Fhit was only identified in one of the above 8 cases. Reduced Fhit expression was observed in 4 among the 30 samples with aberrant transcripts. The remaining 4 cases with reduced Fhit did not show aberrant transcripts. Moreover, Fhit expression was not reduced in the 5 cases where normal transcripts were absent or barely detected. Reduced Fhit expression was observed in only one patient displaying LOH at FHIT with no aberrant transcripts. The remaining six cases with reduced Fhit expression did not display LOH at the FHIT locus. We observed no association between LOH and abnormal FHIT transcripts, abnormal FHIT transcripts and abnormal Fhit expression, or, LOH and abnormal Fhit expression. Moreover, the extents of Fhit were not associated with clinical parameters, such as age, sex, Child-Pugh classification, histological grade of tumor, presence of tumor capsule, AFP, tumor size, microscopic invasion, tumor recurrence, or survival (P > 0.05).
To investigate the association between p53 mutations and genetic changes in the FHIT gene in the pathogenesis of HCC, direct sequencing analysis was performed. p53 mutations were identified in tumors of 9 of the 48 patients (Table 2), but not in the surrounding liver tissues. However, none of the cases displayed G to T transversion at p53 codon 249.
| Mutation sites | n | Mutation sites | n |
| Exon 5 | Exon 8 | ||
| Arg(CGC)158Leu(CTC) | 1 | Arg(CGT)273Cys(TGT) | 1 |
| Exon 6 | Exon 7 | ||
| His(CAT)193Tyr(TAT) | 1 | Arg(TAC)236Cys(TGC) | 1 |
| Ser(AGT)215Arg(AGG) | 1 | Arg(TAC)236Asp(GAC) | 1 |
| Tyr(TAT)220Cys(TGT) | 1 | Met(ATG)246Leu(TTG) | 1 |
| Met(ATG)246Val(GTG) | 1 |
Common fragile sites are prone to genomic alterations in cancer, and the majority of cancers exhibit alterations at fragile sites[26]. The FHIT gene located at one of the most common fragile sites is altered in a variety of tumor cell lines, as well as, in premalignant and malignant lesions of primary tumors, in line with its role as a tumor suppressor gene whose loss or inactivation may contribute to cancer development or malignant progression[1315–20]. Moreover, several studies have been reported results for chromosome 3p rearrangements, decreased or absent FHIT mRNA expression, intragenic deletions and absence of protein expression, in HCC cell lines and primary HCC[1315–192122].
Aberrant FHIT transcripts have been identified in 39%-70% of HCC cases[1315–18]. These transcripts are generated by exon skipping, use of alternative 5’ and 3’ splice sites, and recognition of cryptic splice sites, resulting in insertions of intronic sequences. Consequently, the patterns of aberrant FHIT transcripts vary, but the majority of transcript splicing occurs within two large introns 4 and 5[15–18]. As the fusion functions coincide exactly with splice sites, aberrant transcripts may represent alternatively spliced products[9]. However, point mutations of the FHIT gene are very rare[1315–17]. In our study, all aberrant FHIT transcripts displayed deletion of exons 5 and 6. Additionally, no point mutations in the FHIT gene were identified in the region of the open reading frame. Aberrant transcripts of the FHIT have been identified in both HCC and surrounding non-cancerous tissues[15–18]. In our study, abnormal sized bands representing aberrant RT-PCR products were detected in 22.9% of the surrounding non-tumorous liver tissues and 56.3% of HCCs. These results suggested that aberrant FHIT transcripts had already accumulated even at the chronic liver diseases, in which persistent viral infection and sequential inflammation have occurred. As alterations of the FHIT gene in non-neoplastic tissues of smokers and ex-smokers are observed, FHIT alteration may be an early change during the preneoplastic phase of hepatocarcinogenesis[15–18]. Gramantieri et al reported that HCC cases with aberrant FHIT transcripts showed a significantly higher relapse rate and shorter recurrence time[14]. However, we observed no correlation between aberrant FHIT transcripts and clinicopathological parameters. Ohta et al detected full-length RT-PCR products in nearly all cases with aberrant transcripts[9]. Most of HCCs also exhibited aberrant and normal-sized transcripts[15–18]. Consistent with previous reports, our result showed that almost all cancerous and non-cancerous liver tissues exhibited normal-sized transcripts. In these cases, the normal transcripts have derived from admixed normal cells[9]. In our study, mRNA was extracted from tissue homogenates and, it was therefore not possible to assess whether or not the same cells produced normal and abnormal messengers. However, Chen et al reported that the normal products were observed in all tumor cell lines[17]. It has been suggested that these normal products are derived from neoplastic cells. Moreover, Schlott et al reported that aberrant transcripts did not differ between malignant, benign proliferating, and normal hepatocytes[16]. That is, formation of aberrant FHIT transcripts appears to be a common feature of benign, non-neoplastic hepatocytes. Alternative splicing definitely occurs in normal human tissues[27]. It is thus possible that the FHIT gene is simply located near to but is not the true target that drives a clonal selection process[17]. The FHIT gene, containing a common fragile region, FRA3B, is susceptible to the breakage caused by physical or chemical carcinogens. Similar effects may lead to a higher frequency of changes in the FHIT gene in chronically damaged liver tissues.
Several mechanisms are associated with dysfunction of the FHIT gene, the major being genomic deletion. These chromosomal deletions are frequently identified in HCC cell lines[13]. However, LOH at FHIT gene in human HCC is only occasionally detected[131517]. Our study revealed that LOH of the FHIT gene was found in only 15% of HCCs. Although a great number of loci should be analyzed to further define any possible small deletions and rearrangements, these results so far suggest that LOH of FHIT gene is an uncommon in hepatocarcinogenesis. Allelic loss at FHIT is occasionally observed either in the presence or absence of aberrant transcripts[15]. In our study, LOH of FHIT gene was observed in 6 of 30 cases with aberrant transcripts and 1 of 18 cases without aberrant transcripts. However, no significant correlations were evident between the expression of aberrant transcripts and LOH of FHI. These finding imply that LOH alone does not completely suppress FHIT expression.
Several studies show that whereas all normal and non-cancerous liver tissues show a strong expression of Fhit, most HCC cell lines and primary HCC express reduced or no Fhit[13192122]. However, our study, reduced Fhit expression was observed in only 8 (17%) of 48 HCCs, and complete loss of Fhit in only one of these cases. Moreover, the extent of Fhit expression was not associated with aberrant transcripts or LOH presence. The data suggests that, in some cases, aberrant splice transcripts are actually transcribed. This hypothesis requires confirmation with a larger, more extended study. Inconsistent with our study, the incidence of Fhit expression was lower in HCC developed in patients chronically infected with HBV and exposed to chemical carcinogens, particularly in Qidong, China[1319]. The exact reasons for the variation in Fhit expression are currently unclear, but may be dependent on different geographic and environmental factors. Hepatitis B virus infection is the most common etiologic factors for the development of HCC in South-East Asian regions, such as China and South Korea. Epidemiological and experimental studies disclose synergistic effects of aflatoxin B1 (AFB1) and HBV on hepatocarcinogenesis[28]. Environmental factors, including AFB1, may also contribute to the alteration of the FHIT gene. The concentration of AFB1 in the environment is high in China, but, low in South Korea[29]. AFB1 is strongly associated with a G to T transversion at codon 249 of the p53 gene[28]. However, in our study, none of the cases displayed this G to T transversion. Loss or reduction of Fhit expression in HCC has been identified in association with altered FHIT transcripts, and LOH at the FHIT locus[13]. In our study, reduced Fhit expression was observed in only 4 of 30 cases with aberrant FHIT transcripts, and the remaining 4 cases displayed reduced Fhit expression did not present aberrant FHIT transcripts. Only one case with LOH at FHIT exhibited reduced Fhit expression. The six other cases with decreased Fhit expression did not show LOH at the FHIT locus. In contrast to previous studies, we observed no associations between Fhit expression and altered transcription or LOH. Loss of Fhit expression correlates with poor outcome in various cancers[1430]. Zhao et al also reported that the expression of Fhit is inversely related to disease progression in HCC[19]. However, we found no correlation between Fhit expression and clinicopathological parameters. Although no interrelationship was evident between Fhit expression and LOH events, it is possible that other mutations, not investigated our study, are responsible for reduced or negative protein expression. Therefore, several genetic or epigenetic factors may potentially contribute to the loss of Fhit expression. One possibility is that inactivation of the FHIT gene results from epigenetic mechanisms, such as hypermethylation of 5'-CpG islands in the promoter region[1931]. In addition, abnormalities in post-transcriptional regulation may also abrogate expression of the FHIT gene. However, our results are preliminary and need to be confirmed in a larger study including more cases and an extended follow-up period.
In conclusion, our data indicate that abnormalities in FHIT gene transcripts occurred frequently in both cancerous and non-cancerous liver tissues. However, a normal-sized transcript without sequence abnormalities was almost always present. The Fhit is under-expressed only in a minor fraction of HCC tissues, while it was strongly expressed in non-cancerous liver tissues. Therefore, our study suggests that FHIT plays a role in relatively few HCC in lower AFB1 exposure area such as, South Korea. It is possible that such a finding was attributable to chance due to the relatively small numbers in our study. Thus, additional studies with more subjects are needed to confirm this finding.
The fragile histidine triad (FHIT) has been reported that this gene is a bona fide tumor suppressor gene. The FHIT gene and its product may be involved in the regulation of cell proliferative and apoptotic process. Down-regulation of FHIT inhibits apoptosis and FHIT has been shown to interact with a number of key proteins involved in cancer including p53
Altered transcripts and allelic loss of the FHIT gene are frequently found in premalignant and malignant lesions of various tumors. Moreover, loss or reduction of FHIT protein (Fhit) expression has been found in most tumors including hepatocellular carcinoma (HCC).
Abnormalities of the FHIT gene transcripts occurred frequently in cancerous and non-cancerous liver tissues. However, a normal-sized transcript without sequence abnormalities was almost always present. Moreover, the Fhit was under-expressed only in a minor fraction of HCC tissues in lower AFB1 exposure area such as, South Korea, while it was strongly expressed in non-cancerous liver tissues. Therefore, none of the cases had a G to T transversion at p53 codon 249.
FHIT behaves as a tumor suppressor gene whose loss or inactivation may contribute to HCC development or malignant progression in patients chronically infected with HBV and exposed to chemical carcinogens, particularly in areas from Qidong, China. In contrast, FHIT plays a role in relatively few HCC in lower AFB1 exposure area such as South Korea.
This is an original study that has a large big amount of work. The authors concluded that FHIT plays a role in relatively few HCC’s in South Korea. This is an interesting study, with sound methodology.
| 1. | Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5-S16. |
| 2. | Lok AS. Prevention of hepatitis B virus-related hepatocellular carcinoma. Gastroenterology. 2004;127:S303-S309. |
| 3. | Cha C, Dematteo RP. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol. 2005;19:25-37. |
| 4. | Thomas MB, Zhu AX. Hepatocellular carcinoma: the need for progress. J Clin Oncol. 2005;23:2892-2899. |
| 5. | Kok K, Osinga J, Carritt B, Davis MB, van der Hout AH, van der Veen AY, Landsvater RM, de Leij LF, Berendsen HH, Postmus PE. Deletion of a DNA sequence at the chromosomal region 3p21 in all major types of lung cancer. Nature. 1987;330:578-581. |
| 6. | Naylor SL, Johnson BE, Minna JD, Sakaguchi AY. Loss of heterozygosity of chromosome 3p markers in small-cell lung cancer. Nature. 1987;329:451-454. |
| 7. | Brauch H, Johnson B, Hovis J, Yano T, Gazdar A, Pettengill OS, Graziano S, Sorenson GD, Poiesz BJ, Minna J. Molecular analysis of the short arm of chromosome 3 in small-cell and non-small-cell carcinoma of the lung. N Engl J Med. 1987;317:1109-1113. |
| 8. | Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H. Allelotype study of primary hepatocellular carcinoma. Cancer Res. 1991;51:89-93. |
| 9. | Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587-597. |
| 10. | Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, Inoue H, Tornielli S, Pilotti S, De Gregorio L, Pastorino U. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell. 1996;85:17-26. |
| 11. | Ji L, Fang B, Yen N, Fong K, Minna JD, Roth JA. Induction of apoptosis and inhibition of tumorigenicity and tumor growth by adenovirus vector-mediated fragile histidine triad (FHIT) gene overexpression. Cancer Res. 1999;59:3333-3339. |
| 12. | Nishizaki M, Sasaki J, Fang B, Atkinson EN, Minna JD, Roth JA, Ji L. Synergistic tumor suppression by coexpression of FHIT and p53 coincides with FHIT-mediated MDM2 inactivation and p53 stabilization in human non-small cell lung cancer cells. Cancer Res. 2004;64:5745-5752. |
| 13. | Yuan BZ, Keck-Waggoner C, Zimonjic DB, Thorgeirsson SS, Popescu NC. Alterations of the FHIT gene in human hepatocellular carcinoma. Cancer Res. 2000;60:1049-1053. |
| 14. | Ishii H, Dumon KR, Vecchione A, Fong LY, Baffa R, Huebner K, Croce CM. Potential cancer therapy with the fragile histidine triad gene: review of the preclinical studies. JAMA. 2001;286:2441-2449. |
| 15. | Gramantieri L, Chieco P, Di Tomaso M, Masi L, Piscaglia F, Brillanti S, Gaiani S, Valgimigli M, Mazziotti A, Bolondi L. Aberrant fragile histidine triad gene transcripts in primary hepatocellular carcinoma and liver cirrhosis. Clin Cancer Res. 1999;5:3468-3475. |
| 16. | Schlott T, Ahrens K, Ruschenburg I, Reimer S, Hartmann H, Droese M. Different gene expression of MDM2, GAGE-1, -2 and FHIT in hepatocellular carcinoma and focal nodular hyperplasia. Br J Cancer. 1999;80:73-78. |
| 17. | Chen YJ, Chen PH, Chang JG. Aberrant FHIT transcripts in hepatocellular carcinomas. Br J Cancer. 1998;77:417-420. |
| 18. | Zekri AR, Bahnassy AA, Hafez M, El-Shehaby AM, Sherif GM, Khaled HM, Zakhary N. Alterations of the fragile histidine triad gene in hepatitis C virus-associated hepatocellular carcinoma. J Gastroenterol Hepatol. 2005;20:87-94. |
| 19. | Zhao P, Song X, Nin YY, Lu YL, Li XH. Loss of fragile histidine triad protein in human hepatocellular carcinoma. World J Gastroenterol. 2003;9:1216-1219. |
| 20. | Zochbauer-Muller S, Fong KM, Maitra A, Lam S, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD. 5' CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581-3585. |
| 21. | Tannapfel A, Anhalt K, Hausermann P, Sommerer F, Benicke M, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. J Pathol. 2003;201:238-249. |
| 22. | Nan KJ, Ruan ZP, Jing Z, Qin HX, Wang HY, Guo H, Xu R. Expression of fragile histidine triad in primary hepatocellular carcinoma and its relation with cell proliferation and apoptosis. World J Gastroenterol. 2005;11:228-231. |
| 23. | Greene FL, Page DL, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M. American Joint Committee on Cancer: AJCC Cancer Staging Manual. 6th ed, New York: Springer 2002; 157-164. |
| 24. | Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329-38. |
| 25. | Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. |
| 26. | Croce CM, Sozzi G, Huebner K. Role of FHIT in human cancer. J Clin Oncol. 1999;17:1618-1624. |
| 27. | Boldog F, Gemmill RM, West J, Robinson M, Robinson L, Li E, Roche J, Todd S, Waggoner B, Lundstrom R. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet. 1997;6:193-203. |
| 28. | Smela ME, Currier SS, Bailey EA, Essigmann JM. The chemistry and biology of aflatoxin B(1): from mutational spectrometry to carcinogenesis. Carcinogenesis. 2001;22:535-545. |
| 29. | Park YM, Kim BS, Tabor E. Precore codon 28 stop mutation in hepatitis B virus from patients with hepatocellular carcinoma. Korean J Intern Med. 1997;12:201-207. |
| 30. | Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zöchbauer-Müller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61:8659-8663. |
| 31. | Tanaka H, Shimada Y, Harada H, Shinoda M, Hatooka S, Imamura M, Ishizaki K. Methylation of the 5' CpG island of the FHIT gene is closely associated with transcriptional inactivation in esophageal squamous cell carcinomas. Cancer Res. 1998;58:3429-3434. |