INTRODUCTION
The new global metabolic profiling techniques or ‘metabolomics’, have made it possible to measure large numbers of different metabolites, and are currently being applied to increase our understanding of the health and disease continuum[1]. High-dimensional lipid analysis technologies (lipidomics) provide an opportunity to measure lipids on a broad scale[2]. The majority of lipid pathways involved in lipid metabolism are known, but new lipid metabolites are being discovered all the time. It is not fully known how different pathways affect individual metabolic health and how changes in the regulation of these pathways can influence major metabolic and inflammatory diseases like diabetes, cardiovascular and inflammatory diseases and obesity[2]. The new analytical capacity of lipidomics as a branch of metabolomics can increase our understanding of lipid biology, improve the characterisation of global lipid profiles and result in the identification of previously unknown changes in lipid metabolism[3].
One study has evaluated the transgenomic metabolic effects of two probiotic lactobacilli in mice[4], but as far as we know, the effects of probiotics on global lipidomic profiles in humans have not been characterised before. Previously, probiotics have been shown to possess immunomodulatory effects in in vitro assays, animal models and clinical trials[56], and their effects have been studied mainly in specific conditions such as allergy[78] and inflammatory diseases[9].
In the present study, we characterised the effect of the probiotic Lactobacillus rhamnosus GG (LGG) on global serum lipidomic profiles and investigated whether the changes in inflammatory variables (CRP, TNF-α and IL-6) are reflected in the lipidomics profiles of healthy adults.
MATERIALS AND METHODS
Subjects
The subjects were healthy adults (n = 26, 14 females, 12 males) with a mean age of 42 years (range 23-55) and a mean BMI of 24 kg/m2 (range 20-30). The subjects were recruited to the study by an advertisement in the Helsinki area. The inclusion criteria were being healthy (no chronic illnesses), taking regular exercise (at least three times per week), and not participating in any other clinical trial. The exclusion criteria were milk allergy (due to the nature of the study products), use of antibiotics during the 2 mo before the study, acute gastrointestinal disorders during the 2 mo before the study, gastrointestinal diseases and related medication, pregnancy, and lactation. Before entering the study, the subjects gave their written informed consent. The study protocol was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa.
Intervention
The present study was a substudy of a randomised, double-blind and placebo-controlled parallel group intervention study investigating the immunomodulatory effects of probiotic bacteria with four treatment groups; placebo, LGG, Bifidobacterium animalis ssp. lactis Bb12 and Propionibacterium freudenreichii ssp. shermanii JS[10]. Only the placebo (n = 15) and LGG (n = 11) groups were included in the present pilot study since the LGG exhibited the best anti-inflammatory potential in the original study. Prior to the intervention period, there was a 3-wk run-in period during which no probiotic-containing products were allowed. For 3 wk thereafter, the subjects consumed either a 250 mL milk-based fruit drink containing LGG bacteria (ATCC 53103, 6.2 × 107 cfu/mL) or a similar placebo drink without probiotic bacteria daily. A list of probiotic-containing products was given to the subjects, and they were asked not to consume any other probiotic-containing products at any point during the study. Otherwise they were allowed to eat freely. Venous blood samples from the antecubital vein were taken at baseline and after the 3-wk intervention. The blood samples were stored at -70°C for global lipidomic analyses.
Inflammatory variables and serum lipids
Serum levels of C-reactive protein (CRP) were measured by a high-sensitivity particle-enhanced immunoturbidimetric CRP assay using a Tina-quant CRP (latex) high-sensitivity reagent and a Roche Hitachi 912 analyser (Roche Diagnostics GmbH, Mannheim, Germany) with a detection limit of 0.04 mg/L. All samples were over the detection limit. Cytokine levels (TNF-α, IL-6) in serum were determined using Quantikine HS, Human TNF-α/TNFSF1A (Catalog Number HSTA00D) and IL-6 (HS600B) immunoassays purchased from R&D Systems (Minneapolis MN, USA) according to the manufacturer’s instructions. The detection limit was 0.5 pg/mL for TNF-α and 0.16 pg/mL for IL-6. Serum total cholesterol, high-density lipoprotein (HDL) cholesterol and triglyceride concentrations were measured with their respective enzymatic kits from Roche Diagnostics using an autoanalyser (Roche/Hitachi 912 Automatic Analyzer). Low-density lipoprotein (LDL) cholesterol concentrations were calculated using Friedewald’s equation[11].
Sample preparation for global lipidomic analysis
An aliquot (10 &mgr;L) of an internal standard mixture containing 11 lipid classes and 0.05 mol/L sodium chloride (10 &mgr;L) was added to the serum samples (10 &mgr;L). The lipids were extracted with chloroform/methanol (2:1, 100 &mgr;L). A standard mixture containing 3 labelled standard lipids was added (10 &mgr;L) to the extracts. The sample order for LC/MS analysis was determined by randomization.
Global lipidomics analysis by UPLC/MS
Lipid extracts were analysed on a Waters Q-Tof Premier mass spectrometer combined with an Acquity Ultra Performance LC™ (UPLC). The column, which was kept at 50°C, was an Acquity UPLCTM BEH C18 10 mm × 50 mm with 1.7 &mgr;m particles. The binary solvent system included (A) water (1% 1 mol/L NH4Ac, 0.1% HCOOH) and (B) LC/MS grade (Rathburn) acetonitrile/isopropanol (5:2, 1% 1 mol/L NH4Ac, 0.1% HCOOH). The gradient started from 65% A/35% B, reached 100% B in 6 min and remained there for the next 7 min. The total run time including a 5 min re-equilibration step was 18 min. The flow rate was 0.200 mL/min and the injected amount 0.75 &mgr;L. The temperature of the sample organiser was set at 10°C.
The lipid profiling was carried out on Waters Q-Tof Premier mass spectrometer using ESI+ mode. The data were collected at mass range of m/z 300-1200 with a scan duration of 0.2 s. The source temperature was set at 120°C, and nitrogen was used as desolvation gas (800 L/h) at 250°C. The voltages of the sampling cone and capillary were 39 V and 3.2 kV, respectively. Reserpine (50 &mgr;g/L) was used as the lock spray reference compound (5 &mgr;L/min; 10 s scan frequency).
Data were processed using MZmine software, version 0.60[12]. Lipids were identified using an internal spectral library or by tandem mass spectrometry using UPLC/MS/MS as described previously[13]. The normalisation of lipidomic data was performed as follows: All monoacyl lipids except cholesterol esters, such as monoacylglycerols and monoacylglycerol-PL, were normalised with GPCho (17:0/0:0); diacyl lipids except ethanolamine PL were normalised with GPCho (17:0/17:0); ceramides with Cer (d18:1/17:0); the diacyl ethanolamine phospholipids with GPEtn (17:0/17:0); and the TG and cholesterol esters with TG (17:0/17:0/17:0). Other (unidentified) molecular species were calibrated with GPCho (17:0/0:0) for a retention time of < 300 s, GPCho (17:0/17:0) for between 300 s and 410 s, and TG (17:0/17:0/17:0) for higher retention times. Only the identified lipid molecular species were included in further data analyses.
Lipid nomenclature
Lipids from the global lipidomic analysis were named according to Lipid Maps (http://www.lipidmaps.org). For example, lysophosphatidylcholine (LysoGPCho) with 16:0 fatty acid chain was named monoacyl-glycerophosphocholine GPCho (16:0/0:0). In cases where the fatty acid composition could not be determined, the total number of carbons and double bonds was marked. For example, a phosphatidylcholine species PCho (16:0/20:4) is represented as GPCho (36:4). However, GPCho (36:4) could also represent other molecular species, for example, GPCho (20:4/16:0) or GPCho (18:2/18:2).
Statistical analysis
Principal component analysis (PCA) and partial least squares discriminant analysis (PLS/DA) were utilised as modelling methods for clustering and discrimination[14]. PLS/DA is a pattern recognition technique that correlates variation in the dataset with class membership. The resulting projection model gives latent variables (LVs) that focus on maximum separation (‘‘discrimination’’). The random subsets cross-validation method[15] and Q2 scores were used to develop the models. The VIP (variable importance in the projection) values[16] were calculated to identify the most important molecular species for the clustering of specific groups. PLS/DA and PCA analyses were performed using Matlab, version 7.2 (Mathworks, Natick, MA, USA) and PLS Toolbox, version 4.0, of the Matlab package (Eigenvector Research, Wenatchee, WA, USA. All other analyses were performed using R statistical language (http://www.r-project.org/).
Comparisons between the levels of selected molecular species were performed using the paired Wilcoxon test. For the PLS/DA analyses as well as paired univariate analyses, the data were first log-transformed for each lipid so that X = log (z2/z1), where z2 was the lipid concentration at 21 d and z1 at baseline. With such transformation, the distribution of data was closer to normal and the within-person changes could better be analysed. Chance detection plotting was used to account for multiple hypothesis testing in univariate comparisons. The chance detection plot described how many lipids show more significant differences at random than those actually observed.
In order to assess whether any of the inflammatory variables were explained by global lipidomic profile data, we regressed global lipidomic profile data on selected inflammatory variables using an elastic net method[17]. The method selects an optimal subset of lipids, based on predictive performance of the regression model using extensive bootstrap-based cross-validation. The model is selected by minimum cross-validation-error criterion, which balances the bias against the variance of the estimates. For these analyses, the data were first log-transformed for each lipid/clinical variable so that X = log (z2/z1).
RESULTS
Serum lipids
The mean (SD) baseline value (mmol/L) for total cholesterol was 5.1 (1.1), for LDL cholesterol 3.1 (1.0), for HDL cholesterol 1.5 (0.4) and for triglycerides 1.0 (0.4) in the placebo group and, in the LGG group, 5.4 (1.2), 3.3 (1.0), 1.5 (0.4) and 1.4 (1.1), respectively. The mean (SD) change (mmol/L) during the 3-wk intervention in total cholesterol was 0.2 (0.5), in LDL cholesterol 0.1 (0.5), in HDL cholesterol 0.1 (0.2) and in triglycerides 0.0 (0.5) in the placebo group and 0.0 (0.4), 0.1 (0.3), 0.0 (0.2) and 0.0 (0.6) in the LGG group, respectively. There were no significant differences in serum lipids during the intervention.
Global lipidomic analysis
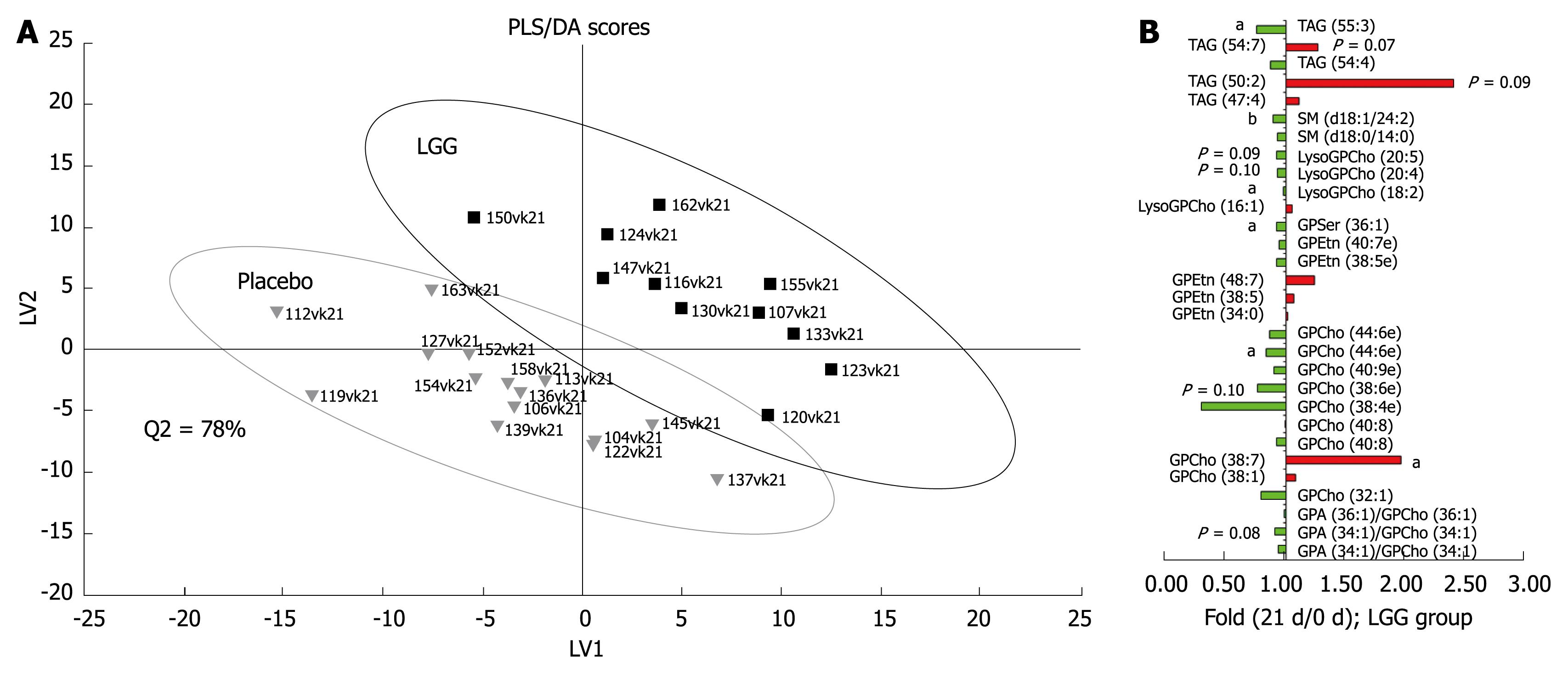
The global lipidomic analysis led to 407 identified lipid species, corresponding to 13 different lipid classes. PCA analysis revealed that no major outliers exist in the data, thus confirming that any changes detected in further analyses would not be due to specific outliers. PLS/DA analysis revealed that the LGG and the placebo groups differed at baseline, and therefore only the within-person changes were utilised in the later statistical analyses. PLS/DA analysis on log-transformed data indicated that the global lipidomic profiles of the groups were separable (Figure 1A). In the LGG group, significant changes (P < 0.05) in lipids were observed during the intervention using paired Wilcoxon test, although when accounting for multiple hypothesis testing by using the chance detection plot, no lipid changes were found to be significant within the 95% confidence interval. However, the VIP analysis revealed some common trends in the lipidomic profile data. Decreased LysoGPCho and sphingomyelins (SM), mainly decreased glycero-phosphatidylcholines (GPCho) and mainly increased triacylglycerols (TAG) were among the most important variables contributing to the separation between the LGG and the placebo groups (Figure 1B).
Figure 1 A: Partial least squares discriminant analysis (PLS/DA) of global serum lipidomic data during the probiotic intervention in healthy adults.
The labels in the picture indicate subject ID numbers; B: Fold changes for the top 30 ranking lipids contributing to the PLS/DA model based on VIP analysis (variable important in the projection) (aP < 0.05, bP < 0.001).
Associations between global lipidomics profiles and inflammatory variables
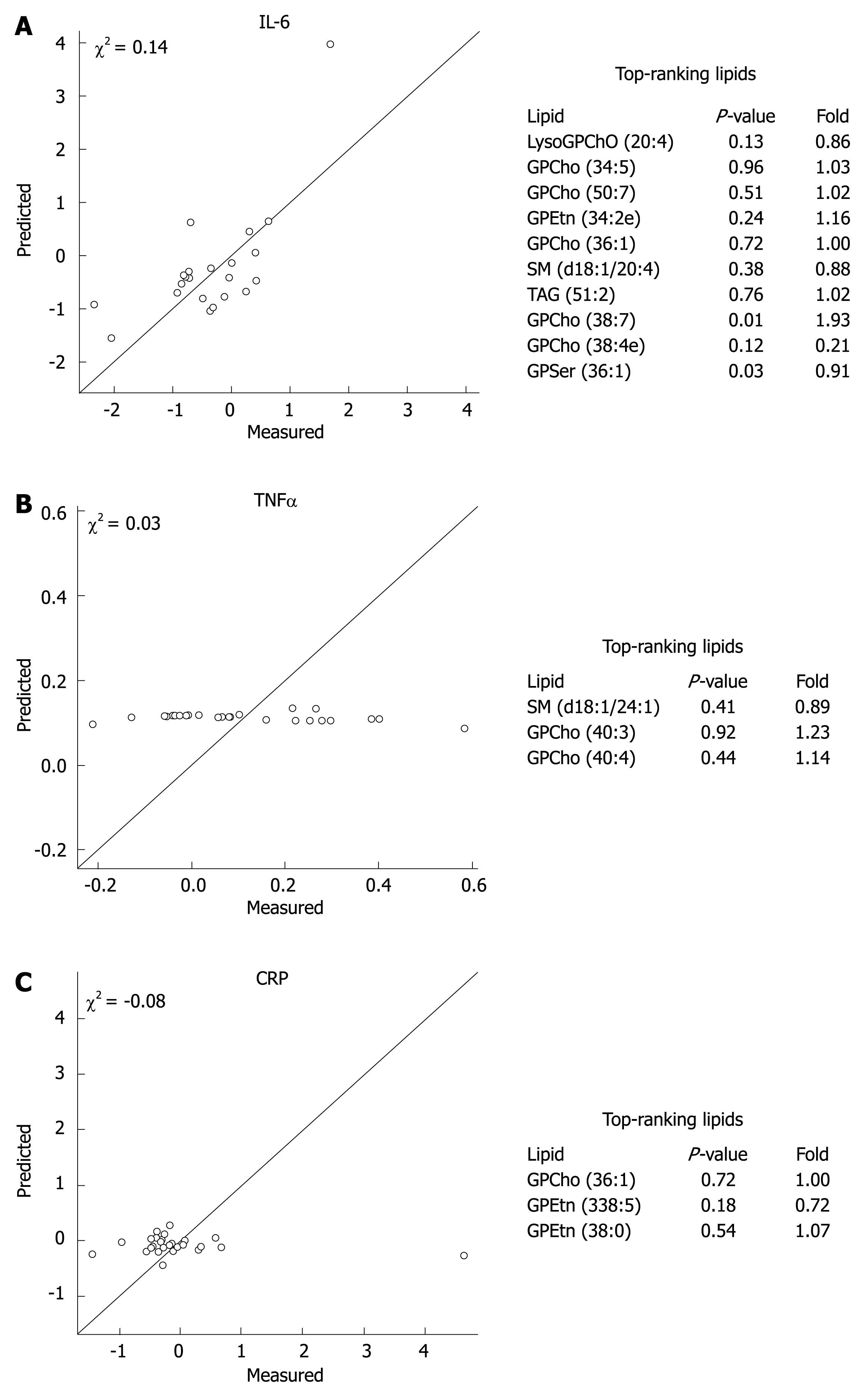
In order to investigate whether the changes in inflammatory variables during the 3-wk intervention were reflected in global lipidomic profiles, we regressed the lipidomic profile data on measured serum TNF-α, IL-6, and CRP values (Figure 2). The results revealed that a reasonably good model based on global lipidomic profiles was found for the proinflammatory cytokine IL-6, while the regression model was poor for CRP and TNF-α. The top-ranking lipid associated with the changes in IL-6 was the proinflammatory LysoGPCho (20:4) (Figure 2).
Figure 2 Cross-validated regression model prediction for IL-6 (A), TNF-α (B) and CRP (C) based on the global lipidomic profile data with the top-ranking lipids explaining the changes in inflammatory variables during the probiotic intervention.
DISCUSSION
This study is the first to apply lipidomic techniques to analyse the global lipidomic profiles of healthy adults after a probiotic intervention. The lipidomic platform has already been applied in multiple studies investigating the pathophysiology of different diseases[18–21]. In the present study, we characterised the effect of probiotic LGG on global serum lipidomic profiles and investigated whether the changes in inflammatory variables (CRP, TNF-α and IL-6) were reflected in global lipidomic profiles in healthy adults. We observed that the probiotic LGG intervention may lead to changes in global lipidomic profiles.
We found a trend towards decreased LysoGPCho after the probiotic LGG intervention. LysoGPCho, derived from phosphatidylcholines, are mediators that affect numerous functions in many types of cells, from proliferation and survival to migration and secretion. They are also involved in oxidative metabolism, angiogenesis, and carcinogenesis[22]. LysoGPCho is a major atherogenic lipid of oxidised LDL[23], and it has been associated with vascular inflammation, endothelial dysfunction and coronary atherosclerosis[24]. LysoGPCho induces an increase in several inflammatory cytokines (IL-1β, IL-6, TNF-α) in human peripheral mononuclear cells (PBMCs)[25]. Therefore, the reduction of LysoGPCho in the present study could be related to our, and previous, results showing a decreased production of TNF-α in PBMCs in healthy adults[1026]. Interestingly, a high LysoGPCho level has been connected also to inflammatory bowel disease (IBD)[2728], impaired mucosal barrier function and increased gut permeability[29–32]. LGG has not been effective in treating Crohn’s disease[3334], but it has been shown to maintain remission in patients with ulcerative colitis[35]. In addition, LGG normalises gut permeability[3637] and enhances mucosal integrity and epithelial cell survival[3839]. Taken together, the decrease in LysoGPCho after LGG intervention observed in the present study may be one of the metabolic events behind the beneficial clinical effects of LGG seen in ulcerative colitis and in normalised gut permeability.
In the present study, we also observed a decrease in SM after the LGG intervention. SM is a major membrane sphingolipid and the precursor of important signalling molecules like ceramide and sphingosine[40]. Recent studies reveal that metabolites of SM are critically important for the initiation and maintenance of diverse aspects of immune cell activation and also function as regulators of inflammatory responses[41–43]. High concentrations of sphingolipids and lipids of the SM/ceramide pathway have been connected to inflammatory processes in the development of atherosclerosis[44] and IBD[4546]. The harmful effects of these lipids may be partly mediated via the production of reactive oxygen species in cells[4247]. As in the case of LysoGPCho, the generation of ceramide by sphingomyelinases from SM and epithelial oxidative stress might contribute to the disturbed barrier function seen in diseases such as IBD[45]. Therefore, the decrease in SM seen after LGG intervention in the present study may also contribute to the beneficial effects on gut barrier function seen in the previous intervention studies with LGG[36–39].
Although we observed some common trends in the global lipidomic profiles after LGG intervention, one should notice that, when accounting for multiple hypothesis testing using the chance detection plot, no lipid changes were found to be statistically significant. This suggests that the study was either underpowered for investigations of global lipidomic profile changes in the described setting, or the observed baseline differences in the global lipidomic profiles dominated over responses to the intervention, masking potential effects of the LGG intervention. One thus cannot exclude the possibility that some of the significant changes were detected by chance. Furthermore, this pilot study was conducted with healthy individuals alone, whereas the effect of LGG intervention on global lipidomic profiles should also be investigated in subjects suffering from inflammatory conditions or disturbed gut barrier function before further conclusions can be drawn.
In conclusion, there are indications that probiotic LGG intervention may lead to changes in global lipidomic profiles reflected in decreased LysoGPCho and SM, mainly decreased GPCho and mainly elevated TAG. These changes may contribute, for example, to the metabolic events behind the beneficial effects of LGG on gut barrier function seen in previous studies. IL-6 was moderately associated with the changes in lipidomic profiles. Lipidomics may provide powerful tools for identifying new biomarkers behind the clinical effects of probiotic intervention trials and for establishing relationships between molecular profiles and other known data from the same individual.
COMMENTS
Background
The new global metabolic profiling technique ‘metabolomics’ has made it possible to measure a large number of metabolites, and is currently being applied to increase the understanding of the health and disease continuum. The new analytical capacity of lipidomics as a branch of metabolomics can increase the understanding of lipid biology, improve the characterisation of global lipid profiles and result in the identification of previously unknown changes in lipid metabolism. Probiotics have been mostly studied in the prevention and treatment of different gastrointestinal diseases and allergy, but the mode of action of probiotics is poorly understood.
Innovations and breakthroughs
This study is the first to apply lipidomic techniques to analyse the global lipidomic profiles of healthy adults after a probiotic intervention. Lipidomic analysis showed that there were decreases in the levels of lysophosphatidylcholines (LysoGPCho), sphingomyelins (SM) and several glycerophosphatidylcholines (GPCho), and increases in triacylglycerols (TAG) in the probiotic LGG group. These changes may contribute, for example, to the metabolic events behind the beneficial effects of LGG on gut barrier function seen in previous studies.
Applications
Metabolomics and lipidomics may help to understand the action mechanisms of different agents, such as probiotics.
Terminology
Lipidomics is a branch of metabolomics which enables identification of lipids in a large scale. Probiotic bacteria are defined as living microorganisms that have beneficial effects on human health.
Peer review
This was an interesting paper and overall it was well written and well-presented. However, this is a small study. One good point is the randomized cohort. The study population was healthy individuals and the results may not be applicable to a population with lipid-related disease states. There were multiple comparisons and one thus cannot exclude the possibility that some of the significant changes were detected by chance.