Published online May 7, 2008. doi: 10.3748/wjg.14.2702
Revised: February 15, 2008
Published online: May 7, 2008
AIM: To clarify alterations of Dickkopfs (Dkks) and Kremen2 (Krm2) in gastrointestinal cancer.
METHODS: We investigated the expression profiles and epigenetic alterations of Dkks and Krm2 genes in gastrointestinal cancer using RT-PCR, tissue microarray analysis, and methylation specific PCR (MSP). Cancer cells were treated with the demethylating agent and/or histone deacetylase inhibitor. WST-8 assays and in vitro invasion assays after treatment with specific siRNA for those genes were performed.
RESULTS: Dkks and Krm2 expression levels were reduced in a certain subset of the gastrointestinal cancer cell lines and cancer tissues. This was correlated with promoter hypermethylation. There were significant correlations between Dkks over-expression levels and beta-catenin over-expression in colorectal cancer. In colorectal cancers with beta-catenin over-expression, Dkk-1 expression levels were significantly lower in those with lymph node metastases than in those without. Down-regulation of Dkks expression by siRNA resulted in a significant increase in cancer cell growth and invasiveness in vitro.
CONCLUSION: Down-regulation of the Dkks associated to promoter hypermethylation appears to be frequently involved in gastrointestinal tumorigenesis.
- Citation: Maehata T, Taniguchi H, Yamamoto H, Nosho K, Adachi Y, Miyamoto N, Miyamoto C, Akutsu N, Yamaoka S, Itoh F. Transcriptional silencing of Dickkopf gene family by CpG island hypermethylation in human gastrointestinal cancer. World J Gastroenterol 2008; 14(17): 2702-2714
- URL: https://www.wjgnet.com/1007-9327/full/v14/i17/2702.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.2702
Wnt proteins influence many aspects such as embryonic development and tumorigenesis, and their activities are regulated by several secreted antagonists[12]. Canonical Wnt signaling via the beta-catenin pathway is transduced by two receptor families. Frizzled proteins and lipoprotein-receptor-related proteins 5 and 6 (LRP5/6) bind Wnts and transmit their signals by stabilizing intracellular beta-catenin. Dickkopfs (Dkks) are secreted antagonists of Wnt signaling and are important for induction of head formation[3]. Dkks contain two distinct cysteine-rich domains in which the positions of 10 cysteine residues are highly conserved between family members. Secreted Dkk-2 and Dkk-4 undergo proteolytic processing that results in cleavage of the second cysteine-rich domain from the full-length protein[4]. Members of the human Dkk-related family differ not only in their structures and expression patterns, but also in their abilities to inhibit Wnt signaling.
The human Dkk-1 (chromosome 10q11.2) gene encodes a powerful inhibitor of the Wnt signaling pathway by binding to and antagonizing LRP5/6[5]. Dkk-1 is a transcriptional target of TCF and p53[45]. Many chemotherapeutic agents that induce DNA adducts significantly enhanced the expression of Dkk-1 in human tumor cell lines, irrespective of their p53 gene status[6]. Dkk-1 expression level was decreased in human colon cancers, mesothelioma, and malignant melanoma, suggesting that Dkk-1 acts as a tumor suppressor gene in these neoplasms[57–9]. The Wnt/beta-catenin pathway was down-regulated by the induction of Dkk-1 expression, a mechanism that is lost in a subset of colon cancers[5]. The Dkk-1 gene was silenced by CpG island promoter hypermethylation in colon cancer cell lines and in 9 (17%) of 54 primary colorectal cancers, especially in advanced Dukes’ C and D colorectal cancers[7]. Dkk-1 showed suppressive effects on tumor growth through beta-catenin-independent non-canonical pathways (i.e., Wnt/JNK pathways) in human mesothelioma[8]. Kremen2 (Krm2) forms a ternary complex with Dkk-1 and LRP6 and induces rapid endocytosis and removal of the Wnt receptor LRP6 from the plasma membrane[10].
Dkk-2 (chromosome 4q25) activates rather than inhibits the Wnt/beta-catenin signaling pathway in Xenopus embryos. Co-transfection of Krm2 blocked the ability of Dkk-2 to activate LRP6 and enhanced inhibition of Wnt/Frizzled signaling in human 293 fibroblasts. Dkk-2 expression was down-regulated in malignant melanoma cell lines and in most tumor samples[9]. Krm2 also cooperates with Dkk-4, but not with Dkk-3, to inhibit Wnt signaling. Krm2 can function as a switch that changes Dkk-2 from an activator to an inhibitor of Wnt/LRP6 signaling[11].
Dkk-3/REIC gene (chromosome 11p15.2) expression level was reported to be decreased in non-small cell lung cancer[1213], renal clear cell carcinoma, urinary bladder cancer, prostate cancer, pancreatic carcinoma, hepatoma, malignant melanoma, and acute lymphocytic leukemia (ALL)[914–16]. Dkk-3 methylation occurred at an early stage in ALL pathogenesis and also influenced the prognosis[15]. Transfection of Dkk-3 into HeLa and Hep3B cells significantly reduced invasion capacity, cell motility, and tumor growth rate in inoculated athymic nude mice[16].
Dkk-4 (chromosome 8p11.2-p11.1) mRNA was expressed in human embryonic stem cells differentiated to an early endodermal cell type, in breast cancer, and in diffuse-type gastric cancer. Dkk-4 is thought to be involved in the negative feedback mechanism of the canonical WNT/beta-catenin signaling pathway[17].
Therefore, altered expression of Dkks and Krm2 appear to play important roles in tumor development and progression. Activated Wnt signal pathway, characterized by the stabilization of beta-catenin, plays an important role in most gastrointestinal cancers. We investigated the expression profiles and epigenetic alterations of the Dkks and Krm2 genes in gastrointestinal cancer in which the Wnt signaling plays an important role.
Human esophageal cancer cell lines (TE 1, 3, 5, 6, 8-11, 14 and 15) were provided by the Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University. Other esophageal cancer (T.T and T.Tn), gastric adenocarcinoma cell lines (AZ521, NUGC3, NUGC4, MKN28, MKN45, MKN74, KATOIII, SNU1, SNU638, HGC27, and GC1Y), colorectal adenocarcinoma cell lines (Colo320DM, DLD1, HCT8, HCT15, HCT116, LoVo, LS123, LS174T, LS180, SKCO1, SW48, SW480, SW620, SW1116, SW1417 and WiDr), hepatocellular carcinoma cell line (CHC4, CHC32, Hep3B, HLF, HuH7 and PLC/PRF), biliary tract adenocarcinoma cell lines (TGBC1TKB and TGBC2TKB), pancreatic adenocarcinoma cell lines (BxPC3, Kp4, MiaPaCa, Panc1 and SuSu86) were purchased from the Japanese Cancer Research Resources Bank (Tokyo, Japan), Riken Cell Bank (Tokyo), or the American Type Culture Collection (Manassas, VA). Cells were cultured in DMEM or RPMI1640 supplemented with 10% fetal bovine serum. Paired specimens of tumor and adjacent non-tumor tissues of the esophagus (n = 10), stomach (n = 24), colorectum (n = 30) and pancreas (n = 7) were purchased from Genomics Collaborative (Laurel, MD). For RT-PCR, tumor cellularity is important. Only specimens containing more than 80% tumor cells were used for analysis.
Semi-quantitative reverse transcriptase-PCR was done as described previously[1819]. Primer sequences were 5’-TCCGAGGAGAAATTGAGGAA-3’ and 5’-C CTGAGGCACAGTCTGATGA-3’ for the Dkk-1 gene, 5’-AGTACCCGCTGCAATAATGG-3’ and 5’-GAAAT GACGAGCACAGCAAA-3’ for the Dkk-2 gene, 5’-CTGGGAGCTAGAGCCTGATG-3’ and 5’-TCA-TACTCATCGGGGACCTC-3’ for the Dkk-3 gene, 5’-AGCTCTGGTCCTGGA CTTCA-3’ and 5’-CAACCCACGACATGTAGCAC-3’ for the Dkk-4 gene, and 5’-ACGC AGCAACACAGCTACAG-3’ and 5’-ATGTGACAGGAGGGGATGTC-3’ for the Krm2 gene. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control of the reaction. All reactions were done at least in duplicate. The levels of gene transcripts were quantified as the ratio of the intensity of the Dkk or Krm2 gene to the intensity of GAPDH. To perform semi-quantitative RT-PCR, the ranges of linear amplification for the Dkk or Krm2 gene and for the GAPDH gene were studied by using standard curves. Underexpression or overexpression was judged when target gene expression in the tumor sample was at least four times lower or higher than that in the corresponding normal sample.
Bisulfite modification of genomic DNA and MSP analysis were performed as described previously[20] using primers corresponding to the Dkk promoter region sequences. Sequences of methylation-specific primers were 5’-CGTTCGTTGGTAGTTTTTATTTCGA-3’ and 5’-GCGACTACCTTTATACCGCGAA-3’ for the Dkk-1 gene, 5’-GAGTAGAGAGAGAGAAAGCGGGAGTTC-3’ and 5’-GTTATCCCCTAACTCACAAAAAACAACG-3’ for the Dkk-2 gene, 5’-CGGTTTTTTTTCGTTTTCGGG-3’ and 5’-CAAACCGCTACATCTCCGCT-3’ for the Dkk-3 gene, 5’-AGAAAAAGTAGTGATAAATAGACGACGT-3’ and 5’-C AACACTATACGTCACCAAAACGAA-3’ for the Dkk-4 gene, and 5’-CGAGGCGGGTAGGAGTTAGTTC-3’ and 5’-CGAAAAAAATCTAACCGAAAACGTT-3’ for the Krm2 gene. Sequences of unmethylation-specific primers were 5’-TGTTTGTTGGTAGTTTTTATTTTG A-3’ and 5’-ACCACAACTACCTTTATACCACAAA-3’ for the Dkk-1 gene, 5’-AGAGAG TAGAGAGAGAGAAAGTGGGAGTTT-3’ and 5’-ATTATCCCCTAACTCACAAAAAAC AACAAA-3’ for the Dkk-2 gene, 5’-TTTTGGTTTTTTTTTGTTTTTGGG-3’ and 5’-CCAA ACCACTACATCTCCACT-3’ for the Dkk-3 gene, 5’-AGAAAAAGTAGTGATAAATAGATGATGT-3’ and 5’-CAACACTATACATCACCAAAACAAA-3’ for the Dkk-4 gene, and 5’-GTGAGGTGGGTAGGAGTTAGTTTGT-3’ and 5’-AAAAAAAATCTAACCAAAAACATT-3’ for the Krm2 gene.
Cancer cells were treated with 2 &mgr;mol/L or 5 &mgr;mol/L of 5-aza-2'-deoxycytidine (5-aza-dC) (Sigma-Aldrich, MO, USA) for 72 h or with 600 nmol/L of trichostatin A (TSA) (ICN Biomedicals) for 24 h. Cells were also treated with 2 &mgr;mol/L of 5-aza-dC for 72 h followed by 600 nmol/L of TSA for an additional 24 h. The timing and sequencing of 5-aza-dC and/or TSA was based on similar preliminary studies as well as published studies[21]. Immediately after completion of treatments, cells were harvested for RNA purification. RT-PCR was performed after 5-aza-dC and/or TSA treatment.
A tissue microarray (SuperBioChips Laboratories, Seoul, Korea) was used for immunohistochemistry. Immunohistochemistry was performed as described previously[2223]. The antibodies used were as follows: goat anti-human Dkk-1 and Dkk-3 (ABCAM, Cambridge, UK), rabbit anti-human Dkk-2 and Krm (ABGENT, San Diego, CA), rabbit anti-human Dkk-4 (Santa Cruz, CA, USA), mouse anti-human Rac (Sigma-Aldrich), rabbit anti-human phospho-Rac (Cell Signaling, MA), rabbit anti-human Ca2+/calmodulin-dependent protein kinaseII (CaMKII) (Santa Cruz), and rabbit anti-human phospho- CaMKII (EPITOMICS, CA, USA). Normal rabbit or mouse immunoglobulins were substituted for each primary antibody as negative controls. The results were analyzed based on the intensity of the expression in the tumors; negative (no staining at high magnification), weak (only visible at high magnification), moderate (readily visible at low magnification) and strong (strikingly positive at low magnification).
Western blot analysis was performed as described previously[18]. The protein signals were visualized by an enhanced chemiluminescence using ECL Western blotting detection reagents (Amersham, IL, USA).
siRNA preparation and in vitro transfection were performed as described previously[22]. Levels of mRNA and protein inhibition were analyzed by RT-PCR and Western blot analysis. siRNA-transfected cells were used for WST-8 and cell invasion assays.
Tumor cell growth was evaluated using the tetrazolium compound WST-8 (Cell Counting Kit-8 Dojindo Laboratories, Kumamoto, Japan) as described previously[22]. Cell viability was determined according to the manufacturer’s instructions.
Assays were performed by the modified Boyden Chamber method as described previously[24]. The results were presented as mean ± SD for each sample.
Data were analyzed by using the computer software package SPSS for Windows 12.0. A P value less than 0.05 was considered statistically significant. The expression level of the Dkks and Krm2 genes was assessed for associations with clinicopathological characteristics using the following statistical tests: Student’s t-test for age; the Mann-Whitney test for histological type, depth of invasion, pathological TNM stage; and the chi-square test or Fisher’s exact test for the remaining parameters. For cell growth and invasion assay, all of the data were analyzed by one-way analysis of variance and the Bonferroni (Dunn) multiple-comparison method.
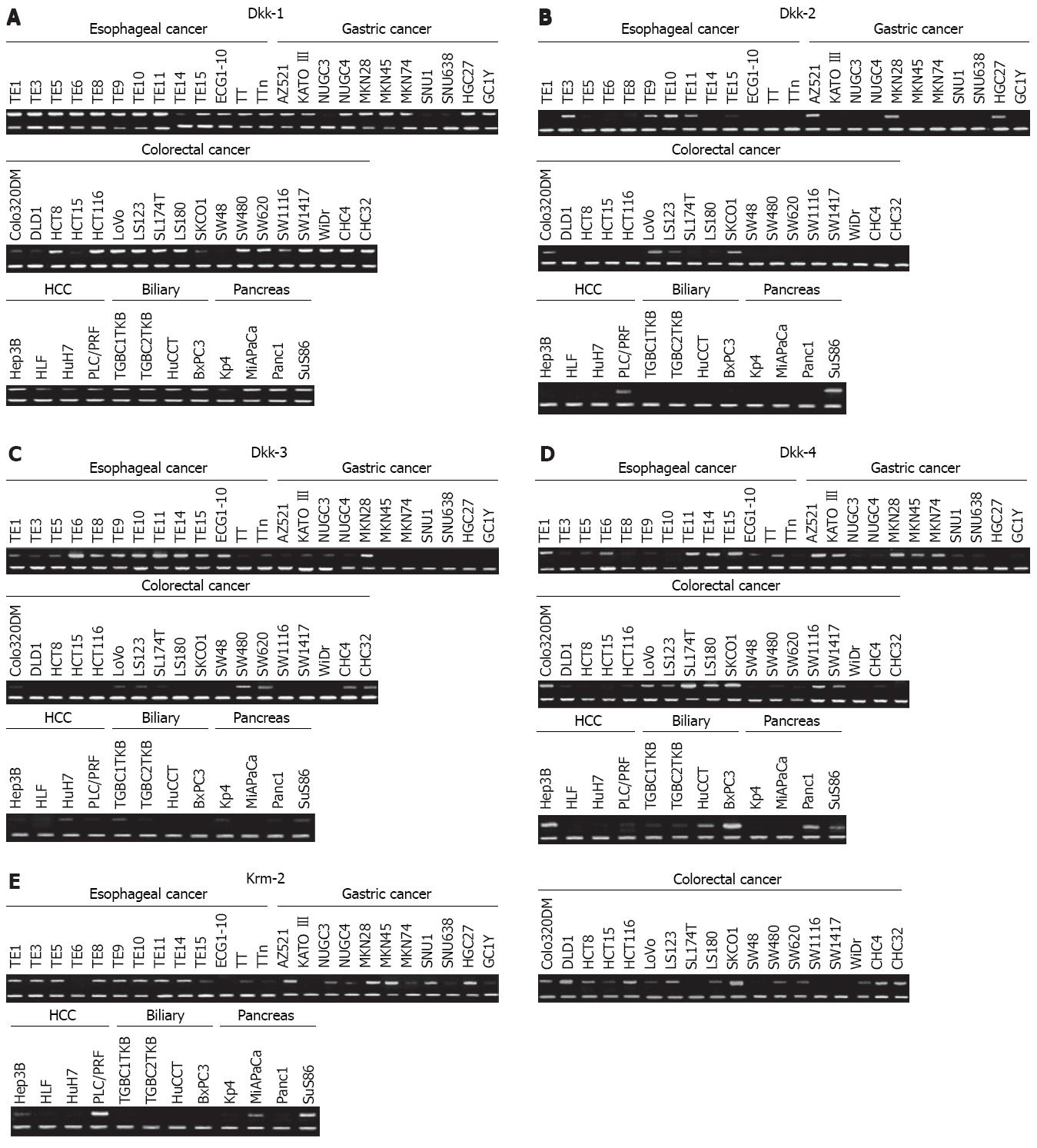
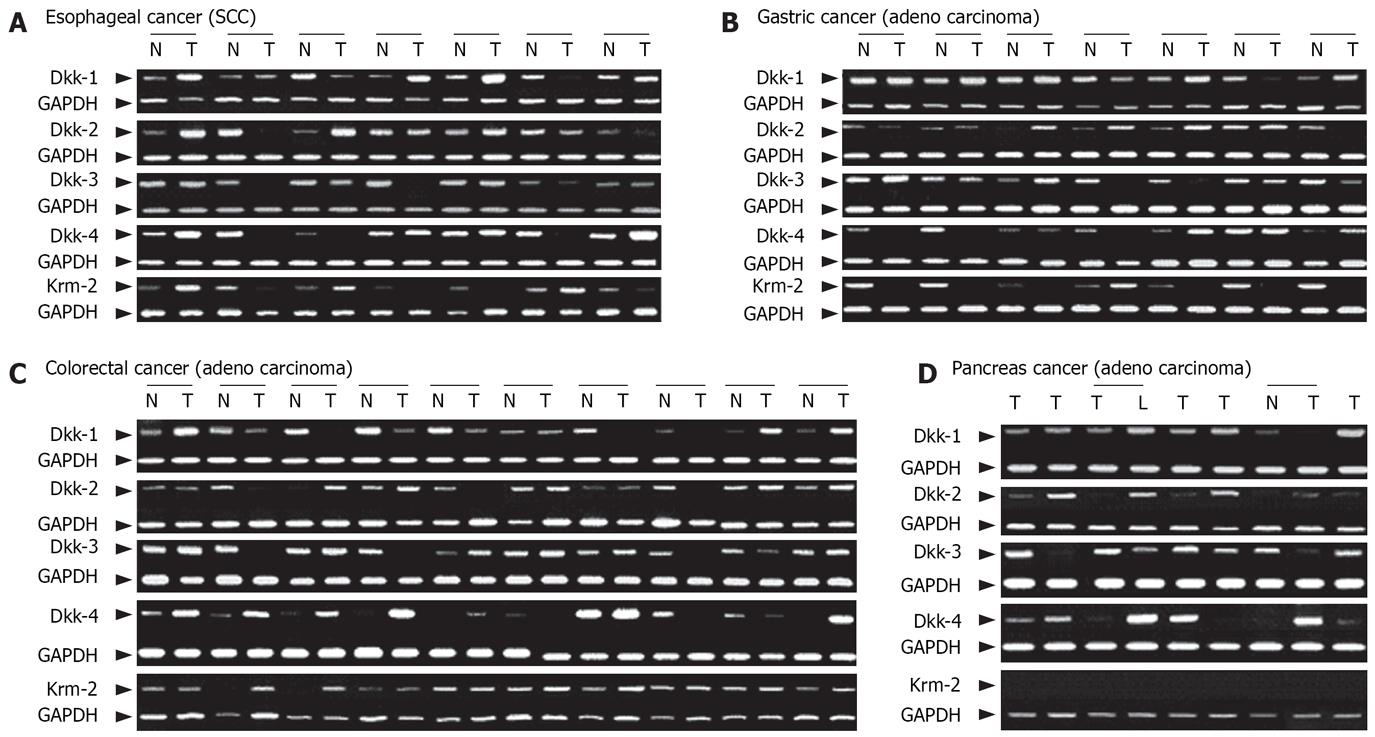
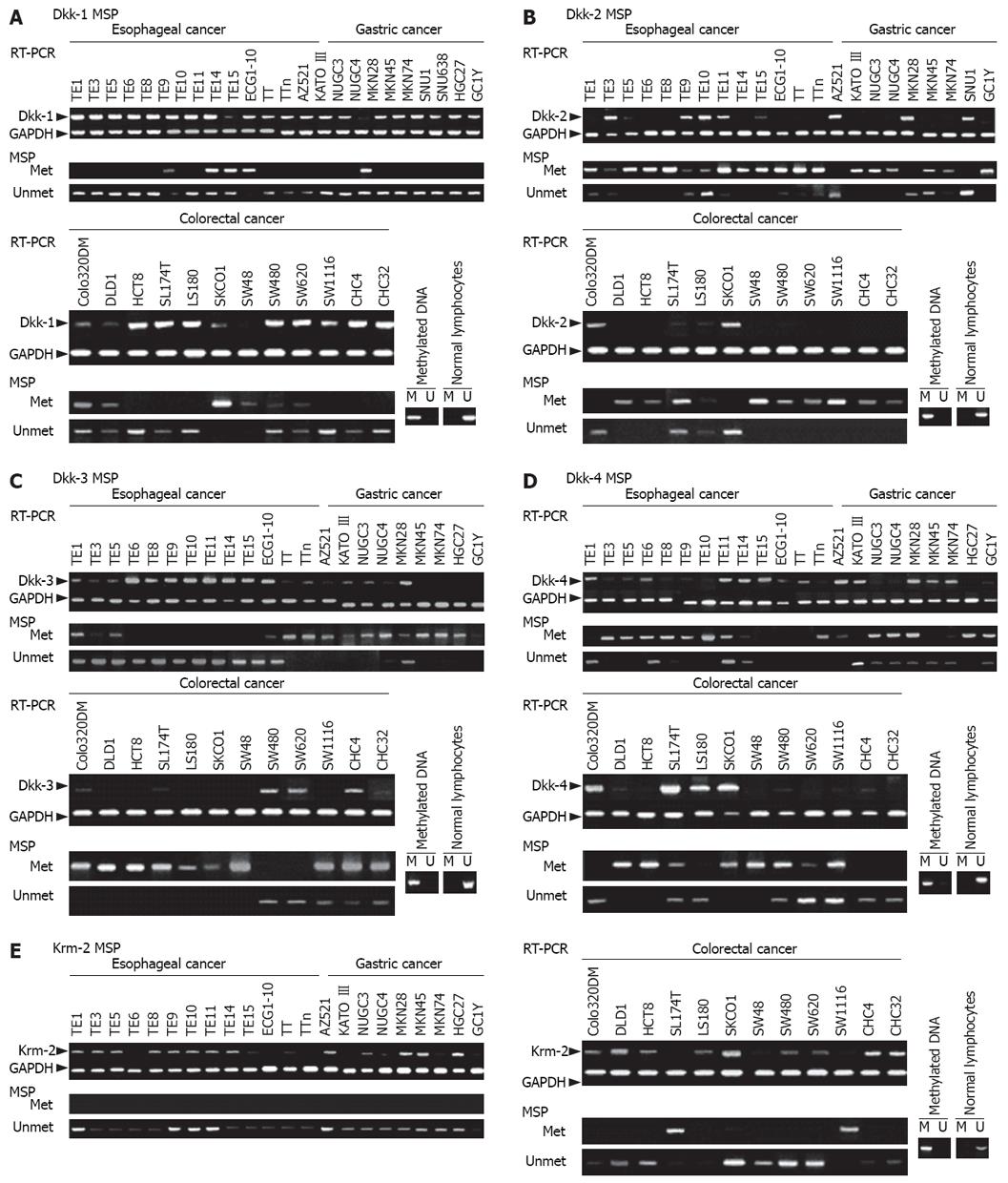
Expression levels of the Dkk genes and Krm2 gene in cancer cell lines and paired normal and cancer tissues were analyzed by using semi-quantitative RT-PCR and immunohistochemistry. The mRNA expression levels of Dkks were reduced in a certain subset of the esophageal, gastric, colorectal, hepatocellular, and pancreatic cancer cell lines (Figure 1). Reduction in the mRNA expression levels of Dkk-1, Dkk-2, Dkk-3, Dkk-4, and Krm2 was observed in 2/10 (20.0%), 3/10 (30.0%), 3/10 (30.0%), 3/10 (30.0%), and 4/10 (40.0%) of esophageal cancer tissues, in 5/24 (20.8%), 2/24 (8.3%), 6/24 (25.0%), 8/24 (33.3%), and 21/24 (87.5%) of gastric cancer tissues, in 6/30 (20.0%), 8/30 (26.7%), 12/30 (40.0%), 4/30 (13.3%), and 0/30 (0%) of colorectal cancer tissues, and in 1/7 (14.3%), 1/7 (14.3%), 2/7 (28.6%), 1/7 (14.3%) and 0/7 (0%; no expression) of pancreatic cancer tissues, respectively (Figure 2 and data not shown). On the other hand, over-expression of Dkks and Krm2 mRNA was also found in these cancer tissues. Dkk-1, Dkk-2, Dkk-3, Dkk-4 and Krm2 mRNA over-expression was observed in 5/10 (50.0%), 3/10 (30.0%), 2/10 (20.0%), 4/10 (40.0%) and 4/10 (40.0%) of esophageal cancer tissues, in 9/24 (37.5%), 16/24 (66.7%), 9/24 (37.5%), 11/24 (45.8%), and 3/24(12.5%) of gastric cancer tissues, in 15/30 (50.0%), 14/30 (46.7%), 7/30 (23.3%), 16/30 (53.3%) and 4/30 (13.3%) of colorectal cancer tissues, and in 3/7 (42.9%), 3/7 (42.9%), 2/7 (28.6%), 3/7 (42.9%) and 0/7 (0%; no expression) of pancreatic cancer tissues, respectively (Figure 2 and some data not shown).
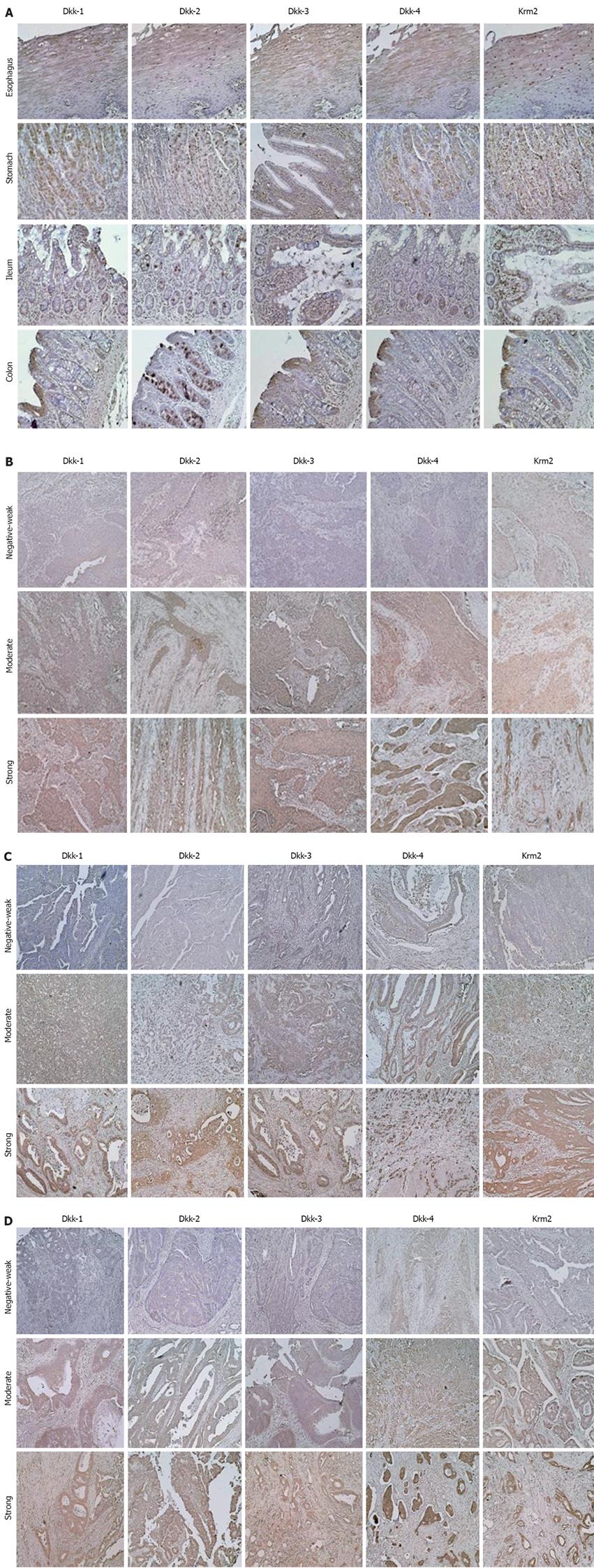
In normal tissues, the squamous epithelium in the esophagus expressed Dkks and Krm2 protein in the more keratinized cells of the epithelium (Figure 3A). Expression levels of Dkk-1, Dkk-2, Dkk-4, and Krm2 protein were strong in secreting cells of the gastric glands. Expression level of Dkk-3 protein was weak in the gastric glands (Figure 3A). In the normal colorectal mucosa, Dkk-1, Dkk-3, Dkk-4, and Krm2 were localized in well-differentiated cells of crypts, whereas Dkk-2 was localized in secreting cells in proliferation layer in crypts (Figure 3A). These results may explain, at least in part, the variable levels of mRNA for these genes in normal tissues. There was no detectable immunoreactivity with the control normal rabbit or mouse immunoglobulins (data not shown).
The protein expression levels of Dkks and Krm2 were reduced in a certain subset of the cancer tissues (Figure 3B-D). The protein expression levels of Dkk-1, Dkk-2, Dkk-3, Dkk-4, and Krm2 were negative or weak in 7/59 (11.9%), 33/59 (55.9%), 18/59 (30.5%), 7/59 (11.9%) and 22/59 (37.3%) of esophageal cancer tissues, in 18/60 (30.0%), 19/60 (31.7%), 20/59 (33.9%), 6/59 (10.2%) and 21/60 (35.0%) of gastric cancer tissues, and in 21/60 (35.0%), 27/60 (45.0%), 27/60 (45.0%), 7/60 (11.7%) and 6/60 (10.0%) of colorectal cancer tissues, respectively. On the other hand, strong staining of Dkk-1, Dkk-2, Dkk-3, Dkk-4, and Krm2 was found in 28/59 (47.5%), 9/59 (15.3%), 10/59 (16.9%), 28/59 (47.5%) and 11/59 (18.6%) of esophageal cancer tissues, in 15/60 (25.0%), 14/60 (23.3%), 22/59 (37.3%), 33/59 (55.9%) and 7/60 (11.7%) of gastric cancer tissues, and in 10/60 (16.7%), 12/60 (20.0%), 6/60 (10.0%), 35/60 (58.3%) and 28/60 (46.7%) of colorectal cancer tissues, respectively (Figure 3B-D and data not shown). Expression levels of Dkks and Krm2 were not correlated with any of the clinicopathological characteristics. However, in colorectal cancers with beta-catenin over-expression, Dkk-1 expression levels were significantly lower in those with lymph node metastasis than in those without lymph node metastasis (P = 0.003).
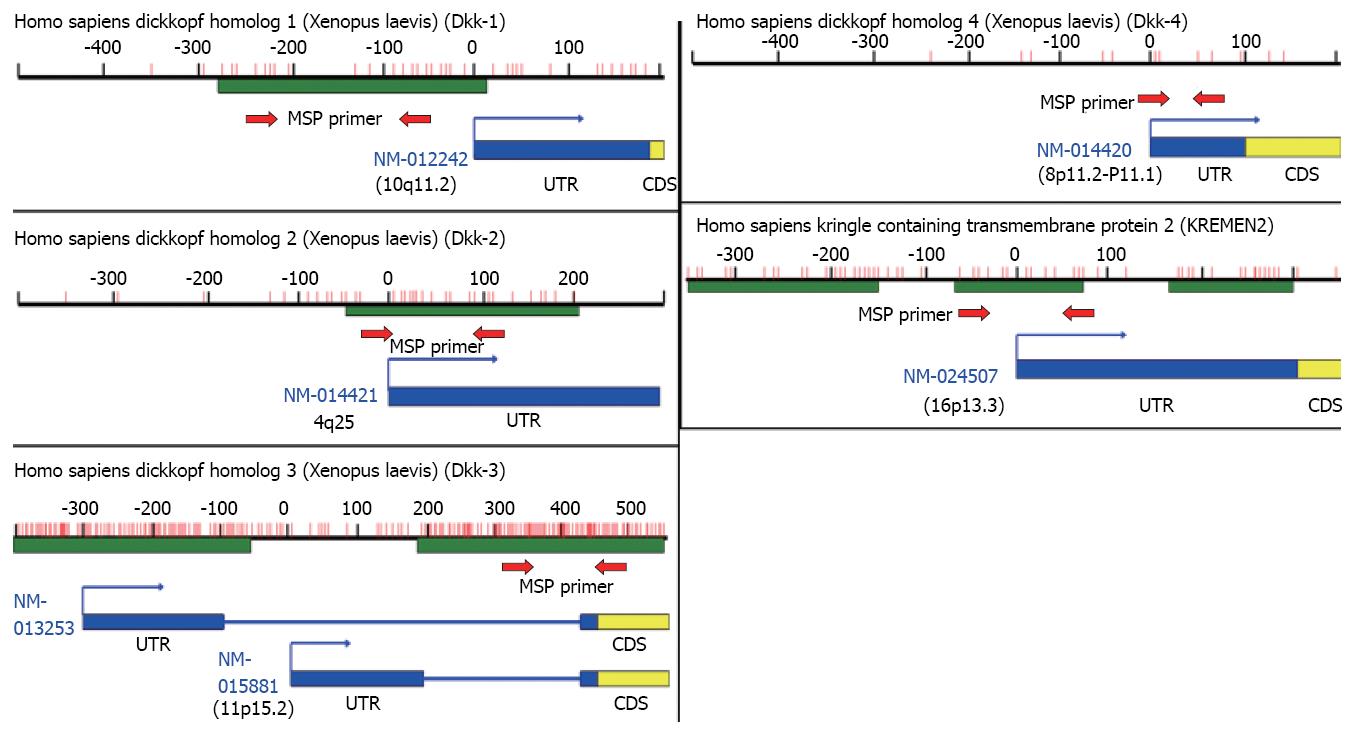
Using Blast and CpG island searcher, we found that Dkk-1, Dkk-2, Dkk-3, and Krm2 genes contain CpG islands at their 5'ends and that Dkk-4 has a few CpGs in the promoter region (Figure 4). Therefore, we analyzed the methylation status of the CpG islands of these genes in cancer cell lines and tissue samples using MSP. Methylation status was significantly associated with silencing of Dkks mRNA expression in cancer cell lines (Figure 5 and data not shown). Methylation status was associated with silencing of Krm2 mRNA expression in only some cancer cell lines, such as LS174T and SW1116 cells (Figure 5E). In cancer tissues, Dkk-1, Dkk-2, Dkk-3, and Dkk-4 were hypermethylated in 4/11 (36.4%), 1/11(9.1%), 1/11 (9.1%) and 3/11 (27.3%) of esophageal cancer tissues, in 2/8 (25.0%), 2/8 (25.0%), 2/8 (25.0%) and 3/8 (37.5%) of gastric cancer tissues, and in 7/20 (35.0%), 13/20 (65.0%), 7/20 (35.0%) and 4/20 (20.0%) of colorectal cancer tissues, respectively (data not shown).
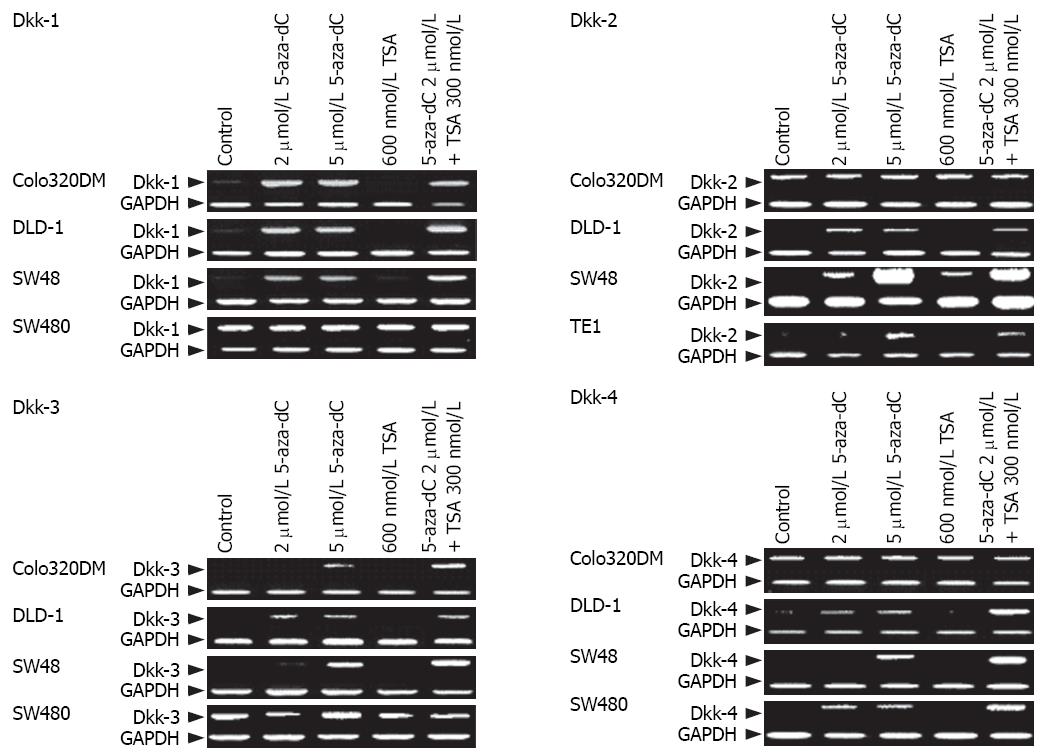
To confirm the role of epigenetic change in silencing of the Dkk genes, cell lines that lacked the Dkk genes expression were treated with 5-aza-dC and/or TSA. Treatment with 5-aza-dC resulted in restoration of Dkk-1 in colo320DM, DLD-1, and SW48 cells, restoration of Dkk-2 expression in DLD-1, SW48, and TE1 cells, restoration of Dkk-3 expression in colo320DM and DLD-1 cells, and restoration of Dkk-4 expression in DLD-1, SW48, and SW480 cells (Figure 6). However, treatment with TSA had no effect.
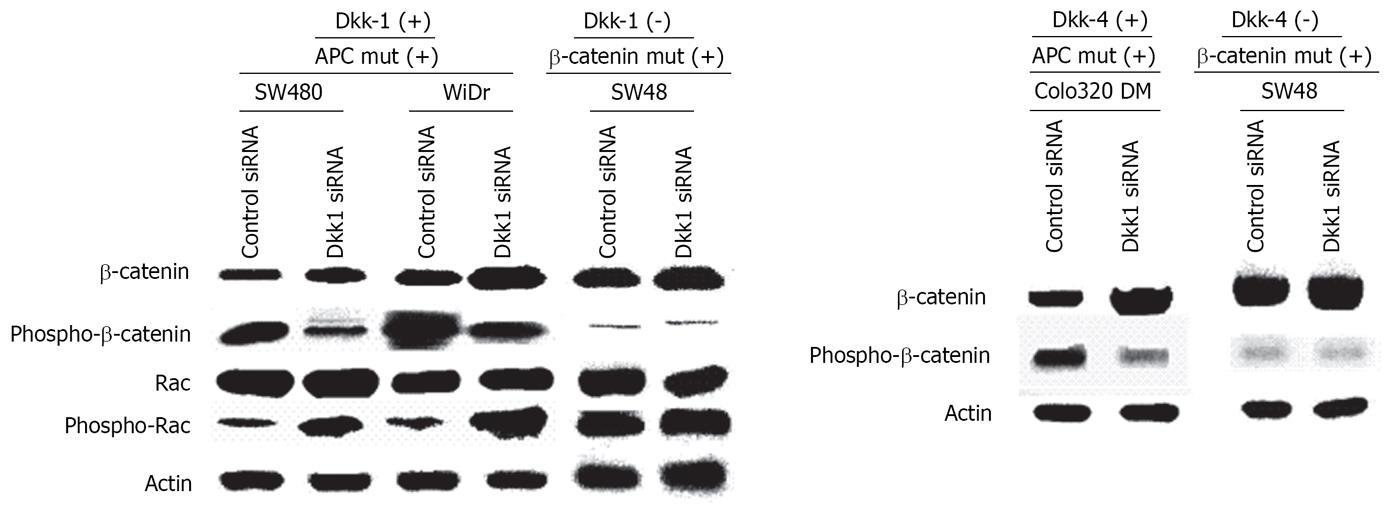
There were some binding motifs of beta-catenin (5’-YCTTTGWW-3’) in the promoter regions of Dkk-1 and Dkk-4. Dkk-2, Dkk-3 and Krm2 did not have TCF-binding motifs in their promoter regions. There were significant correlations between levels of Dkk-1, Dkk-3, Dkk-4 and Krm2 and those of beta-catenin expression in colorectal cancer tissues in tissue microarray data obtained by immunohistochemistry. Dkk-1 (r = 0.500, P < 0.0001), Dkk-3 (r = 0.326, P = 0.0130), Dkk-4 (r = 0.480, P = 0.0003) and Krm2 (r = 0.454, P = 0.0005) expression levels showed significant correlations with beta-catenin over-expression. However, no correlation was found between expression levels of these molecules and beta-catenin over-expression in gastric or esophageal cancer tissues (data not shown). We then analyzed expression levels of beta-catenin and phosphorylated beta-catenin in Dkk1 or Dkk4-specific siRNA-treated colon cancer cells with APC mutation (SW480, WiDr, and colo320DM cells) and SW48 cells with beta-catenin mutation using Western blot analysis. SW480 and WiDr cells treated with Dkk1-specific siRNA showed higher levels of beta-catenin expression and lower levels of phosphorylated beta-catenin compared with those of the control siRNA-treated cells (Figure 7). On the other hand, Dkk-1-negative SW48 cells treated with Dkk1-specific siRNA showed no difference compared with the control cells. Colo320DM cells treated with Dkk4-specific siRNA showed higher levels of beta-catenin expression and lower levels of phosphorylated beta-catenin compared with those of the control siRNA-treated cells (Figure 7). On the other hand, Dkk4-negative SW48 cells treated with Dkk4-specific siRNA showed no difference compared with the control cells.
Next, we examined expression of Rac and phosphory-lated Rac/CDC42, which Wnt4/5A/11 activates through Dvl, and CaMKII and phosphorylated CaMKII to investigate the effects on Wnt-PCP and Wnt-Ca2+ signal transduction. Expression of Rac, phospho-Rac, CaMKII, and phospho-CaMKII was not correlated with any of the clinicopathological features. Rac activation ratio ((phospho-Rac) level-(Rac) level/(Rac)level) was negatively correlated with Dkk-1 in colorectal cancer (r = -0.366, P = 0.0053) and with Dkk-2 (r = -0.290, P = 0.0299) and Dkk-3 (r = -0.3880, P = 0.0037) in esophageal cancer, but not in gastric cancer. On the other hand, CaMKII activation ratio was positively correlated with Dkk-2 (r = 0.285, P = 0.0314) and Krm2 (r = 0.3640, P = 0.0075) in esophageal cancer, but not in colorectal cancer or gastric cancer. We analyzed Rac expression level and the phosphorylation status of Rac in cells treated with Dkk1-specific siRNA using Western blot analysis. SW480 and WiDr cells treated with Dkk1-specific siRNA showed similar expression levels of Rac and higher levels of phosphorylated Rac compared with those of cells treated with the control siRNA (Figure 7). On the other hand, Dkk-1-negative SW48 cells treated with Dkk1-specific siRNA showed no difference compared with the control cells.
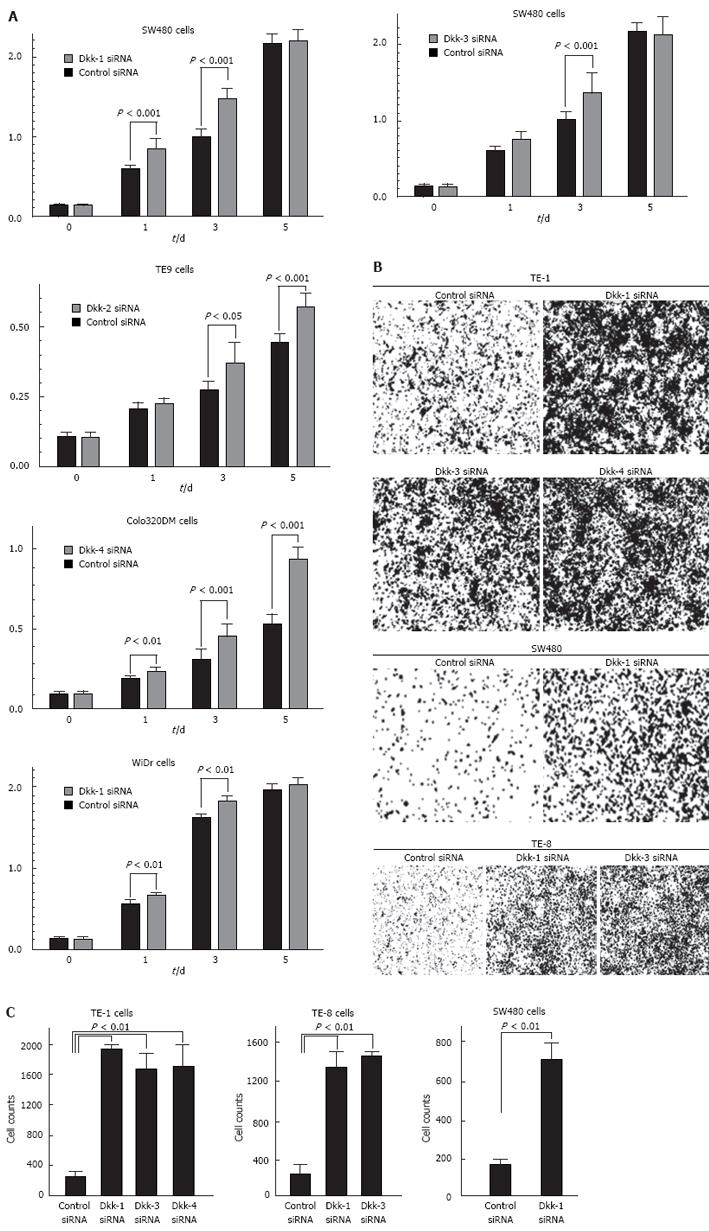
WST-8 assays and in vitro invasion assays after treatment with specific siRNA for the Dkk genes were performed to assess the role of the expression of Dkks in cancer cell growth and invasiveness. Transfection with siRNA resulted in over 80% inhibition of mRNA and protein expression (data not shown). Transfection with Dkk1-specific siRNA enhanced the growth of SW480 and WiDr cells compared with control siRNA-transfected counterparts (Figure 8A; P < 0.01). Transfection with Dkk2-, Dkk3- and Dkk4-specific siRNA enhanced the growth of TE9, SW480 and colo320DM cells compared with control siRNA-transfected counterparts, respectively (Figure 8A; P < 0.05, P < 0.001 and P < 0.001). Transfection with Dkk-1, Dkk-3, or Dkk-4-specific siRNA enhanced the invasiveness of TE-1 cells compared with control siRNA-transfected counterparts (Figure 8B and CP < 0.01). Transfection with Dkk-1 or Dkk-3-specific siRNA enhanced the invasiveness of TE-8 cells compared with control siRNA-transfected counterparts (Figure 8B and C, P < 0.01). Transfection with Dkk-1-specific siRNA enhanced the invasiveness of SW480 cells compared with control siRNA-transfected counterparts (Figure 8B and C, P < 0.01).
In the present study, we investigated the expression profiles and epigenetic alterations of the Dkks and Krm2 genes in gastrointestinal cancer. Dkks and Krm2 expression levels were reduced in a certain subset of gastrointestinal cancer cell lines and cancer tissues. Methylation of Dkk-1, Dkk-2, Dkk-3 and Dkk-4 was significantly correlated with down-regulation of expression in gastrointestinal cancer cell lines and cancer tissues. Moreover, Dkk-1, Dkk-2, Dkk-3 and Dkk-4 mRNA expression was restored by treatment with the DNA-demethylating agent. These results suggest that promoter hypermethylation is an important mechanism of silencing of Dkk family in gastrointestinal cancer. Although there were only a few CpGs in the Dkk-4 promoter region, these CpG sites in cancer tissues were more methylated than in normal tissues. Similarly, the expression of rat placental lactogen-1 (rPL-I) has been reported to be controlled by DNA methylation although the gene has only 17 CpG sites in the 5’-flanking region[25].
Over-expression of Dkk-1, Dkk-2, Dkk-3 and Dkk-4 was also found in a subset of gastrointestinal cancer tissues. This is not a surprising result, because of the following reasons. There are binding motifs of beta-catenin in the promoter regions of Dkk-1 and Dkk-4. Multiple beta-catenin/TCF4 sites in the Dkk-1 gene promoter have been reported to contribute to Dkk-1 activation, thus initiating a negative feedback loop[5]. In fact, Dkk-1 mRNA levels have been increased in a subset of colorectal cancer tissues compared to normal tissues[5]. In our study, Dkk-1, Dkk-3, Dkk-4 and Krm2 expression was correlated with beta-catenin over-expression in colorectal cancer tissues. Moreover, knockdown of Dkk-1 or Dkk-4 up-regulated levels of beta-catenin expression and down-regulated levels of phosphorylated beta-catenin. Therefore, Dkks and Krm2 could be directly and/or indirectly induced by Wnt signals, at least in part, as components of negative feedback loops; but, this mechanism may be lost or abolished in a certain subset of colorectal cancers by promoter hypermethylation. On the other hand, no correlation was found between expression levels of Dkks and Krm2 and beta-catenin over-expression in gastric or esophageal cancer tissues. Further analysis is required to clarify the mechanism of over-expression of Dkks and Krm2 in a subset of gastric and esophageal cancers.
Next, we examined correlations of altered expression of Dkks and Krm2 with expression of the Wnt non-canonical pathway genes. It has been reported that the Wnt canonical pathway activation stabilizes beta-catenin and that the non-canonical pathway activates Rho, Rac, JNK, and PKC or activates CaMKII[26]. The TAK1-NLK MAPK cascade is activated by the non-canonical Wnt-5A/Ca2+ pathway and antagonizes canonical Wnt/beta-catenin signaling[2728]. In the present study, Rac activation was negatively correlated with Dkk-1 expression in colorectal cancer and with Dkk-2 and Dkk-3 in esophageal cancer. These results suggest that Dkk-1 down-regulation in colorectal cancer and Dkk-2 or Dkk-3 down-regulation in esophageal cancer play a role in Rac activation. Moreover, knockdown of Dkk-1 using Dkk-1-specific siRNA induced Rac phosphorylation. On the other hand, CaMKII activation showed a weak positive correlation with Dkk-2 and Krm2 in esophageal cancer. Although further investigation is required, these results suggest additional effects of Dkks and Krm2 on the Wnt non-canonical pathway.
In colorectal cancers with beta-catenin over-expression, Dkk-1 expression levels were significantly lower in those with lymph node metastasis than in those without lymph node metastasis. Dkk-1 promoter has been reported to be hypermethylated in advanced Dukes’ C and Dukes’ D colorectal cancers[7]. Transfection of Dkk-3 has been reported to reduce invasion of osteosarcoma and malignant melanoma[9]. We further revealed that down-regulation of Dkks expression by siRNA resulted in a significant increase in esophageal and colon cancer cell growth and invasion in vitro. Thus, down-regulation of Dkks may contribute to the more aggressive phenotype of esophageal and colorectal cancer cells.
Our results indicate that promoter hypermethylation is an important mechanism of silencing of Dkk family in gastrointestinal cancer. Suzuki et al[2930] reported epigenetic inactivation of secreted frizzled-related proteins (sFRPs) in colorectal and gastric cancer. We reported silencing of the Wnt inhibitory factor-1 gene due to promoter hypermethylation in gastrointestinal tumors[18]. Thus, CpG island promoter hypermethylation is a common mechanism of the inactivation of extracellular Wnt antagonists in gastrointestinal cancer. Activated Wnt signal pathway, characterized by the stabilization of beta-catenin, plays an important role in most gastrointestinal cancers. Modulation of the Wnt pathway, through reversal of extracellular Wnt antagonists silencing by demethylating agents, may be a potential target for treatment and/or prevention of gastrointestinal cancer.
Dickkopfs (Dkks) are secreted antagonists of Wnt signaling pathway. Activated Wnt signal pathway, characterized by the stabilization of beta-catenin, plays an important role in most gastrointestinal cancers. Extracellular Wnt antagonists such as secreted frizzled-related proteins (SFRPs) and WIF-1 is frequently inactivated in gastrointestinal cancer. Thus, Dkks may be also inactivated in gastrointestinal cancer.
Epigenetic transcriptional silencing of tumor suppressor genes plays a key role in gastrointestinal cancer and becomes one of the most important research areas. CpG island promoter hypermethylation is a common mechanism for the inactivation of extracellular Wnt antagonists such as SFRPs and WIF-1 in gastrointestinal cancer.
The expression profiles and epigenetic alterations of Dkks (Dkk1, Dkk2, Dkk3 and Dkk4) and Kremen2 (Krm2) genes were systematically analyzed in many gastrointestinal cancer cell lines and tissues by using RT-PCR, tissue microarray analysis, and methylation specific PCR (MSP).
Our study demonstrated that down-regulation of the Dkk gene family and Krm2 gene associated to promoter hypermethylation is frequently involved in gastrointestinal tumorigenesis. Hypermethylated Dkks could be a marker for screening gastrointestinal cancer. Modulation of the Wnt pathway, through reversal of Dkks and/or Krm2 gene silencing by demethylating agents, may be a potential target for treatment and/or prevention of gastrointestinal cancer.
This paper studied alterations of Dkks and Krm2 in gastrointestinal cancer. The authors showed that down-regulation of the Dkks and Krm2 associated to promoter hypermethylation is frequently involved in gastrointestinal tumorigenesis. Hypermethylated Dkks could be a marker for screening gastrointestinal cancer and reactivation of Dkks and/or Krm2 gene could be a promising strategy for treatment and/or prevention of gastrointestinal cancer.
| 1. | Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627-2634. |
| 2. | Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042-4045. |
| 3. | Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357-362. |
| 4. | Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301-313. |
| 5. | Gonzalez-Sancho JM, Aguilera O, Garcia JM, Pendas-Franco N, Pena C, Cal S, Garcia de Herreros A, Bonilla F, Munoz A. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098-1103. |
| 6. | Wang J, Shou J, Chen X. Dickkopf-1, an inhibitor of the Wnt signaling pathway, is induced by p53. Oncogene. 2000;19:1843-1848. |
| 7. | Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, Garcia JM, Munoz A, Esteller M, Gonzalez-Sancho JM. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116-4121. |
| 8. | Lee AY, He B, You L, Xu Z, Mazieres J, Reguart N, Mikami I, Batra S, Jablons DM. Dickkopf-1 antagonizes Wnt signaling independent of beta-catenin in human mesothelioma. Biochem Biophys Res Commun. 2004;323:1246-1250. |
| 9. | Kuphal S, Lodermeyer S, Bataille F, Schuierer M, Hoang BH, Bosserhoff AK. Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene. 2006;25:5027-5036. |
| 10. | Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664-667. |
| 11. | Mao B, Niehrs C. Kremen2 modulates Dickkopf2 activity during Wnt/LRP6 signaling. Gene. 2003;302:179-183. |
| 12. | Nozaki I, Tsuji T, Iijima O, Ohmura Y, Andou A, Miyazaki M, Shimizu N, Namba M. Reduced expression of REIC/Dkk-3 gene in non-small cell lung cancer. Int J Oncol. 2001;19:117-121. |
| 13. | Tsuji T, Nozaki I, Miyazaki M, Sakaguchi M, Pu H, Hamazaki Y, Iijima O, Namba M. Antiproliferative activity of REIC/Dkk-3 and its significant down-regulation in non-small-cell lung carcinomas. Biochem Biophys Res Commun. 2001;289:257-263. |
| 14. | Kobayashi K, Ouchida M, Tsuji T, Hanafusa H, Miyazaki M, Namba M, Shimizu N, Shimizu K. Reduced expression of the REIC/Dkk-3 gene by promoter-hypermethylation in human tumor cells. Gene. 2002;282:151-158. |
| 15. | Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Barrios M, Andreu EJ, Prosper F, Heiniger A, Torres A. Transcriptional silencing of the Dickkopfs-3 (Dkk-3) gene by CpG hypermethylation in acute lymphoblastic leukaemia. Br J Cancer. 2004;91:707-713. |
| 16. | Hsieh SY, Hsieh PS, Chiu CT, Chen WY. Dickkopf-3/REIC functions as a suppressor gene of tumor growth. Oncogene. 2004;23:9183-9189. |
| 17. | Katoh Y, Katoh M. Comparative genomics on DKK2 and DKK4 orthologs. Int J Mol Med. 2005;16:477-481. |
| 18. | Taniguchi H, Yamamoto H, Hirata T, Miyamoto N, Oki M, Nosho K, Adachi Y, Endo T, Imai K, Shinomura Y. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946-7952. |
| 19. | Hirata T, Yamamoto H, Taniguchi H, Horiuchi S, Oki M, Adachi Y, Imai K, Shinomura Y. Characterization of the immune escape phenotype of human gastric cancers with and without high-frequency microsatellite instability. J Pathol. 2007;211:516-523. |
| 20. | Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. |
| 21. | Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103-107. |
| 22. | Taniguchi H, Yamamoto H, Akutsu N, Nosho K, Adachi Y, Imai K, Shinomura Y. Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic structure in human gastrointestinal cancer. J Pathol. 2007;213:131-139. |
| 23. | Miyamoto N, Yamamoto H, Taniguchi H, Miyamoto C, Oki M, Adachi Y, Imai K, Shinomura Y. Differential expression of angiogenesis-related genes in human gastric cancers with and those without high-frequency microsatellite instability. Cancer Lett. 2007;254:42-53. |
| 24. | Yamamoto H, Vinitketkumnuen A, Adachi Y, Taniguchi H, Hirata T, Miyamoto N, Nosho K, Imsumran A, Fujita M, Hosokawa M. Association of matrilysin-2 (MMP-26) expression with tumor progression and activation of MMP-9 in esophageal squamous cell carcinoma. Carcinogenesis. 2004;25:2353-2360. |
| 25. | Cho JH, Kimura H, Minami T, Ohgane J, Hattori N, Tanaka S, Shiota K. DNA methylation regulates placental lactogen I gene expression. Endocrinology. 2001;142:3389-3396. |
| 27. | Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798-802. |
| 28. | Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J, Matsumoto K. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:131-139. |