Published online Feb 28, 2008. doi: 10.3748/wjg.14.1204
Revised: December 12, 2007
Published online: February 28, 2008
AIM: To study the molecular mechanism of laterally spreading tumor (LST), a cell line [Laterally Spreading Tumor-Rectum 1 (LST-R1)] was derived and the characteristics of this cell line were investigated.
METHODS: A new cell line (LST-R1) originated from laterally spreading tumor was established. Properties of the cell line were characterized using scanning and transmission electron microscopy, immunohistochemistry method, cytogenetic analysis and nude mice xenograft experiments. In vitro invasion assay, cDNA microarray and Western blotting were used to compare the difference between the LST-R1 and other colorectal cancer cell lines derived from prudent colon cancer.
RESULTS: Our study demonstrated that both epithelial special antigen (ESA) and cytokeratin-20 (CK20) were expressed in LST-R1. The cells presented microvilli and tight junction with large nuclei. The karyotypic analysis showed hyperdiploid features with structural chromosome aberrations. The in vivo tumorigenicity was also demonstrated in nude mice xenograft experiments. The invasion assay suggested this cell line has a higher invasive ability. cDNA microarray and Western blotting show the loss of the expression of E-cadherin in LST-R1 cells.
CONCLUSION: We established and characterized a colorectal cancer cell line, LST-R1 and LST-R1 has an obvious malignant tendency, which maybe partially attributed to the changes of the expression of some adhesion molecules, such as E-cadherin. It is also a versatile tool for exploring the original and progressive mechanisms of laterally spreading tumor and the early colon cancer genesis.
- Citation: Wang XY, Lai ZS, Yeung CM, Wang JD, Deng W, Li HY, Han YJ, Kung HF, Jiang B, Lin MCM. Establishment and characterization of a new cell line derived from human colorectal laterally spreading tumor. World J Gastroenterol 2008; 14(8): 1204-1211
- URL: https://www.wjgnet.com/1007-9327/full/v14/i8/1204.htm
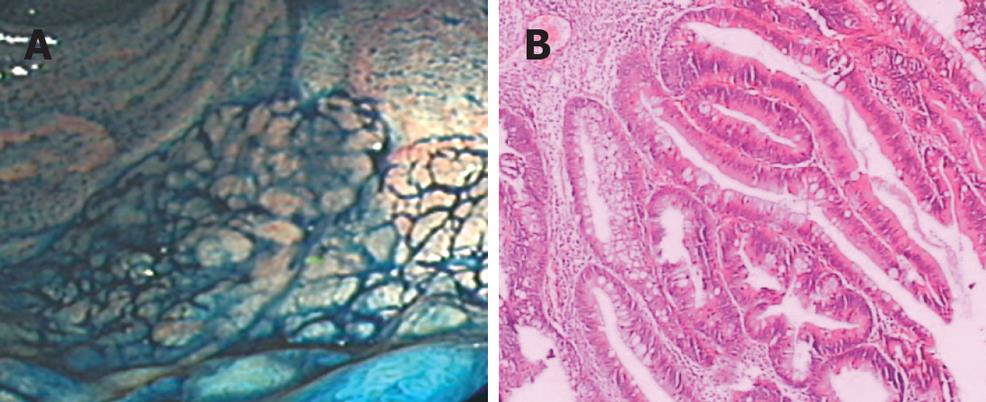
- DOI: https://dx.doi.org/10.3748/wjg.14.1204
Two routes have been described that can lead to colon carcinogenesis: one is the adenoma-carcinoma model, which is the main carcinogenetic pathway of prudent colorectal tumor and has been widely accepted in the west. However, an increasing number of superficial or flat colorectal tumors have been reported, particular in Japan[12]. The superficial tumors are considered to have a distinct clinicopathologic, genetic feature and behave a different carcinogenetic pathway, called de novo pathway[3–6]. Laterally spreading tumor (LST) is a unique subtype of superficial colorectal tumor, which was first reported by Kudo S in 1993[78] and many names such as granular cluster type, nodule-aggregating tumor, creeping tumor, superficial spreading (epithelial) tumor, flower-bed-like lesion had been used. In 1998, it was defined as tumors originated from the colorectal mucous membranes and mainly extends laterally rather than vertically with its diameter greater than 1 cm (Figure 1A)[9–12]. It has a tendency to affect the rectum, sigmoid colon, and caecum. In some studies, it has been reported that the carcinogenesis rate of LST is 8.64%-52.5%[13–15]. Moreover, our previous study has also shown that the incidence of LST is not low in China[16]. Flat appearance makes it difficult to detect it and previous study has shown that laterally spreading tumors have a higher malignant potential than conventional adenomas[21718]. To study the distinctive biological behavior and the underlying biological mechanisms responsible for LST cancer progression will certainly help improving the detection rate of LST and eventually lead to the decreasing morbidity of colon cancer.
Invasion is an important step in the intricate process leading to the formation of cancer and has close relationship with the adhesion ability of the cancer cells. To investigate the higher malignant tendency of colorectal superficial tumor, we established a stable cell line derived from LST and assessed its invasion ability by in vitro invasion assay. Here we show that the invasion ability of LST is higher than SW480 and lovo cells, which is originated from prudent colon cancer. To elucidate the reason for this difference, we have developed a cDNA microarray, representing 18 000 cDNA clusters to profile the gene expression patterns in Laterally Spreading Tumor-Rectum 1 (LST-R1), SW480, lovo cell lines and found that many genes associated with adhesion showed a different expression profile. Our data suggest that LST-R1cells have some distinct characteristics comparing with SW480 and lovo cells. Further investigations on the cells should enhance our understanding on the distinct biology of LST.
Laterally spreading tumor cells were derived from a rectal LST of 59-year-old Chinese woman. Magnifying endoscope showed a flat granular tumor with nodus (about 70 mm × 60 mm) in rectum, 3 cm far from anus. Examination of the biopsy specimen revealed that it has the characteristic morphology of a villous adenoma accompanied by moderate sever atypical hyperplasia (Figure 1B).
Tissue specimens were obtained by endoscopic partial mucosal resection (EPMR), and were transferred to a transport medium containing five folds penicillin, streptomycin (Invitrogen, Carlsbad, CA) and amphotercin B. The specimens were vibrated for about ten minutes to get rid of filth and washed five times with transport medium. They were then trimmed to remove fat and connective tissue, minced into pieces in a sterile culture dish, and subsequently plated in a 25 cm2 flask. Cells were incubated in a 37°C incubator with 5% CO2. RPMI1640 medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA)was added to the cells four hours later and media was changed every 2-3 d. Clonal growth was observed after 27 d of culture, and colonies were identified and subcultured. After 2 passages, the cells grew rapidly and were polymorphic with a few fibroblasts. To purify the cells, they were inoculated in 96-well plates and were cultured separately to get rid of fibroblasts. Finally, the polymorphic cells (more than 90%) were designated as LST-R1.
HCT116, lovo, SW480 and colo205 colon tumor cell lines were commercially obtained from the American Type Culture Collection and maintained in RPMI 1640 medium supplemented with 100 U/mL penicillin,100 &mgr;g/mL streptomycin, 10% fetal bovine serum in 75 cm2 tissue culture flasks at 37°C in a 5% CO2 environment.
Cells were grown on chamber slides and stained for CK20 and ESA expression with a specific antibody (Beijing ZhongShan Biology technology Ltd, Beijing, China).
LST cells grown on slides were trypsinized and centrifuged. After fixed in 2.5% glutaraldehyde and postfixed in 1% osmium, they were dehydrated with acetone. The sample was divided into two parts: one half was added with acetas, dried on critical point of CO2 and observed under scanning electron microscope, another half was embedded in paraffin and cut into thin pieces with a thickness of about 700 mm, stained with uranyl acetate and citrate, and observed under transmission electron microscope.
Metaphase cells were obtained by treatment of the cultured cells with Colcemid (Gibco, Grand Island, NY, USA) at a final concentration of 0.03 &mgr;g/mL for 3 h. The harvested cells were treated with 0.8% sodium citrate for 15 min at 37°C and fixed in 3:1 methanol/acetic acid. Metaphase chromosome spreading was performed as previously described[19]. The slide with metaphase cells was aged at room temperature (R.T.) for 5-7 s prior to SKY. The slide was treated with DNase-free RNase solution (0.1 g/L) at 37°C for 1 h, and briefly rinsed in distilled water. After washing in 2 × SSC for 10 min at R.T, the slide was treated with proteinase K (0.5 &mgr;g/mL) solution for 10 min at 37°C and washed in 2 × SSC at R.T. for 10 min. The slide was then fixed in 1% paraformaldehyde and washed in 2 × SSC at R.T. for 10 min each, followed by dehydration in 70%, 85%, and 95% ethanol at R.T. for 2 min. The slide was placed in 70% formamide/2 × SSC at 70°C for 4 min and dehydrated in 70%, 85%, and 95% ethanol for 2 min each. Other protocols including SKY probe (Applied Spectral Imaging, Migdal Ha’Emek, Israel) denaturation, hybridization and detection were done according to the recommendations of SKY probe manufacturer. SKY image capturing and karyotyping were performed using the SkyVision Imaging System equipped with a Zeiss Axioplan 2 fluorescence microscope. Recommendations were followed in karyotyping descriptions[20].
To determine the population doubling time and mitotic index of LST-R1 cells, the cells were suspended at a concentration of 1 × 105 per mL and 100 &mgr;L suspension was cultured in each well of 96 well plates. The cell growth was monitored after 12 h, 24 h, 36 h and 48 h by 3-(4, 5-dimethylthiazol- 2-yl)-2, 5-diphenyltetrazolium bromide (MTT, the final concentration = 0.5 g/L) assay. The value indicated the mean of three independent experiments.
In vivo tumorigenicity studies were performed using 5-wk-old athymic nude mice obtained from the Animal Center of the First Military Medical University (Guangzhou, China). LST-R1 cells at passage 51 were trypsinized, washed twice with serum-free RPIM1640 medium, and resuspended in serum-free RPMI-1640 (1 × 106 per mL). A final volume of 0.2 mL was injected per site on the back of 8 athymic nude mice. Tumor growth was measured weekly using calipers. The tumor area was calculated according to the following formula: tumor area = π× (width/2 × length/2). When the tumors grew to a certain extent (about 6 mm × 4 mm), tissue was removed and fixed in neutral buffered formalin for histological examination. Some cells from the xenografts were re-established in vitro to compare their morphology with that of the original cell line.
The invasion assay was performed by using 24-well BD BioCoat™ Matrigel™ Invasion Chamber with 8-&mgr;m polycarbonated filters (Becton Dickinson, Bedford, MA), as described by Albini et al[21]. 1 × 104 cells were seeded on Matrigel invasion chamber plates and cultured in routine medium. Cells were incubated for 22 h at 37°C in a humidified incubator with 5% CO2. Nonmigratory cells on the upper surface of the filter were removed by wiping with a cotton swab. Invasive cells that penetrated through pores and migrated to the underside of the membrane were stained with Giemsa solution after fixation with 4% formaldehyde in PBS. The cell number was counted under microscopic vision.
Total RNA was extracted using standard Trizol RNA isolation protocol (Life Technologies, Inc., Grand Island, NY). The poly (A) mRNA was isolated from the total RNA using a poly (dT) resin (Qiagen, Hilden, Germany). Nylon membrane-based cDNA microarrays were prepared by using Spotter (BioRobotics, Cambridge, U.K.) containing clones from the present work and other tissue resources and the internal controls. The procedures for probe preparation, hybridization, washing, scanning, and signal intensity normalization of the spots were performed also using a previously described method[22]. The differential expression was considered as significant between LST and SW480, lovo cell lines when the ratio of signals between the same spots on different membranes was greater than 2.
Western blotting analysis was assessed according to the protocol described before[23]. In brief, cells were washed with ice-cold PBS twice and lysed with ice-cold lysis buffer. Protein samples (15 &mgr;g) were separated by SDS/PAGE (12% acrylamide gel) using a Bio-Rad Mini-Protean III system (45 mA for about 2 h). Proteins were transferred to PVDF membranes and the membranes were blocked for 1 h at room temperature with TBST. Blots were then incubated at room temperature with primary antibodies (1:500 dilution) and then primary antibodies were removed and the blots were extensively washed with TBS/Tween 20 min for three times. Blots were then incubated for 1 h at room temperature with the secondary antibodies (1:2000 dilution). Then blots were extensively washed as above for 1 h and developed using the Enhanced Chemiluminescence detection system and quantified using the GeneTools systems.
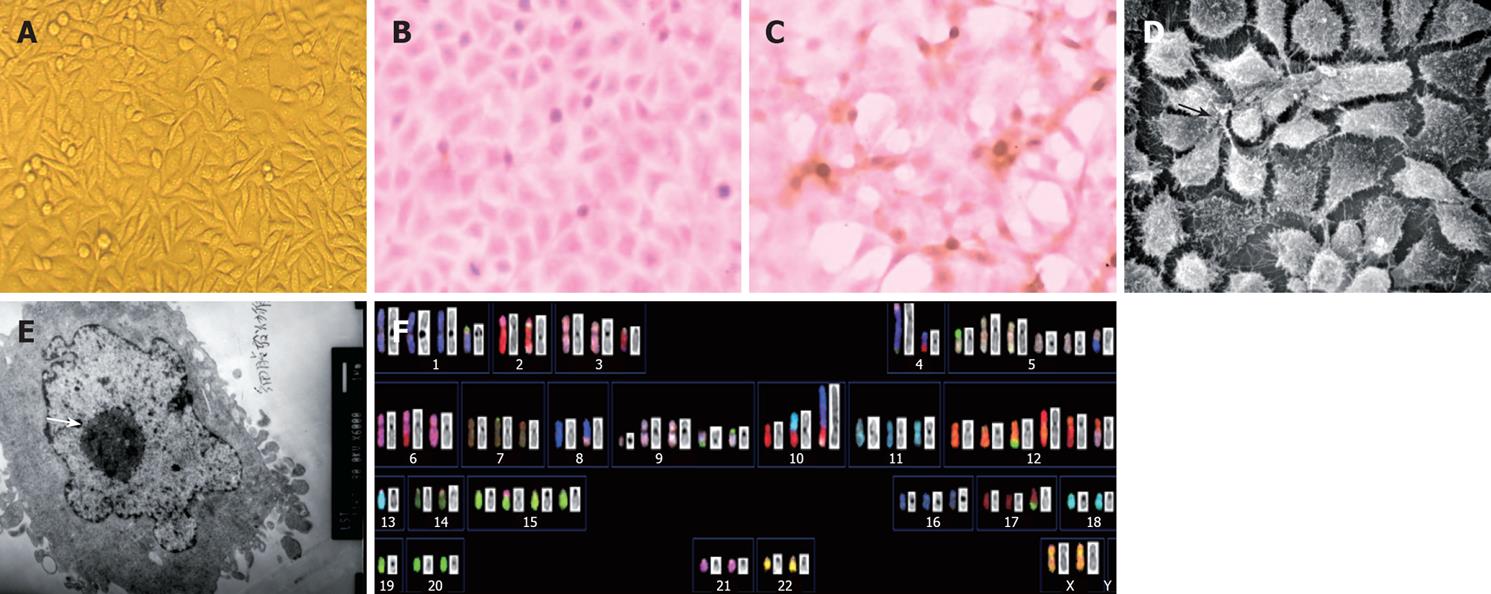
When established in vitro, LST-R1 cells grew as attached cells with spindle or polygonal morphology. 90% of the individual cells have 1 or 2 heterogenous nuclei with 1 or 2 prominent nucleoli (Figure 2A). To confirm the epithelial origin of this cell line, expression of ESA and CK20 were determined by immunocytochemistry. The positive expression of ESA and CK20 staining revealed that the cells are of epithelial origin (Figure 2B and C). Under the scanning electron microscope, microvilli and tight junction were observed. There are fewer organelles (such as Golgi apparatus and endoplasmic reticulum) and more secretory granules were observed (Figure 2D and E).
| 64-66,XX,+der(1)t(1;3)(p11;?)t(3;19)(?;?)[21],der(2)t(2;6)(p11;q23)[30],der(2)t(2;22)(p21;?)t(3;22)(?;?)t(2;3)(?;?)[9],der(2)t(2;22)(p21;?)t(3;22)(?;?)t(2;3)(?;?)t(2;3)(q24;?)[21],-2[30],del(3)(p24)[30],-3[9],der(3)del(p12)t(2;3)(q21;q11)[21],-3[9],der(4)t(4;10)(q11;p11)[30],der(4)dup(14)(q22q31)t(4;14)(q?;q?)[9],der(4)t(3;4)(p21;p16)dup(14)(q22q31)t(4;14)(q?;q?)[21],-4[30],der(5;8)(p10;p10)[30],+i(5)(p10)×2[30],+der(5;20)(p10;q10)del(5)(q31)[30],der(6)t(2:6)(p13;q23)[30],der(7)t(7;15)(p22;q?)[30],der(7)t(7;17)(p13;?)[29],der(8)(3;8)(?;q22)[30],der(9)t(9;19)(p11;?)[30],+der(9)t(9;19)(q11;?)[30],+r(9)(p13q13)[29],der(10;13)(q10;q10)[30],der(10)t(4;10)(p11;11)t(4;8)(q31;q13)t(3;10)(p23;q23)[30],der(12)t(3:12)(q24;q11)[30],der(12)t(5;12)(?;p11)[30],+der(12)t(2;12)(q22;q11)[30],+der(12)t(2;12)(p?;p11)t(2;12)(q24;q?)[30],+der(12)t(12;20)(q24;?)[30],-13[30],-14[30],+der(15)t(6;15)(?;p11)[30],der(17)t(15:17)(q25;q24)[30],-18[30],-19[30],-19[30],-21[30],der(22)t(3;22)(?;p13)[9],-22[30][cp30] |
Karyotypic analysis was performed on the LST-R1 cell line at passage 111 by spectral karyotypic technique. The composite karyotypes were summarized in Table 1. Basically, two subclones were identified. Although there were some karyotypic differences between the two subclones, the majority of chromosome aberrations existed in all 30 metaphases analyzed, indicating that the cells were derived from a single progenitor aberrant cell. Chromosome numbers differed in a narrow range (from 64 to 66). A representative karyotype of LST-R1 cells with 66 chromosomes is shown in Figure 2 F.
Generally, the cells were triploid with complex structural chromosome aberrations and a number of numerical abnormalities.
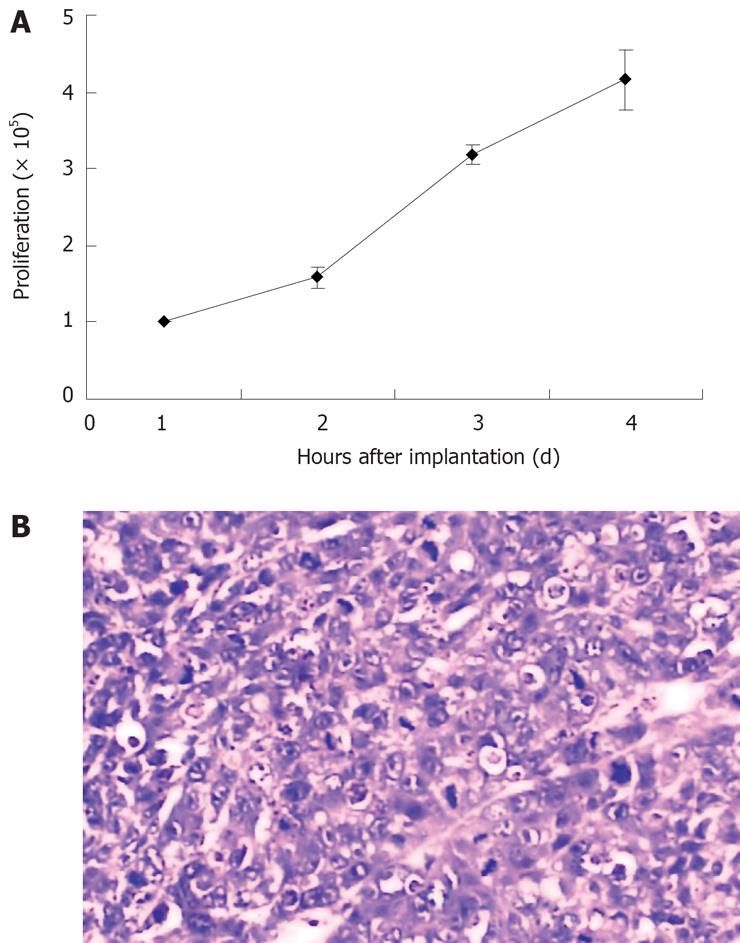
LST-R1 cells were passaged for more than 110 times. In medium containing 10% FBS, the cells grew as monolayers with varying degrees of attachment. They grew rapidly with large nuclei. The population doubling time was 36 h, and the mitotic index arrived at the peak value at the fourth day in a 25-cm2 flask (Figure 3A).
The in vivo tumorigenicity of LST-R1 cells was studied using athymic nude mice xenograft. Tumor formation was observed 15 s after inoculation, and tumors grew in all of the 8 mice. Tumor area progressively increased and reached an average of 3 cm2 in size 28 s after inoculation. The histopathology of the xenograft tumor showed that it was a poorly differentiated adenocarcinoma (Figure 3B). Cells cultured from the xenograft tumor (using the same protocol as previously described) had the same properties as the original cell line.
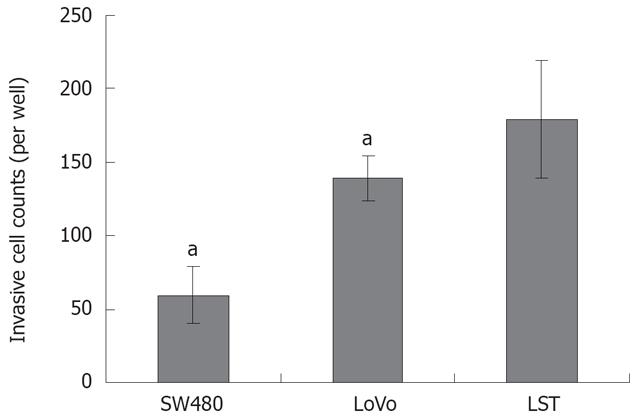
In vitro invasion assay revealed that invasive activity increased significantly in LST-R1 comparing with the other two cell lines. Twenty-four hours later, the numbers of LST-R1 cells invading the membrane were significantly higher (179.3 ± 40 cells/well) than those of lovo (139.3 ± 15.1 cells/well) and SW480 (59.3 ± 19.1 cells/well) cells (Figure 4).
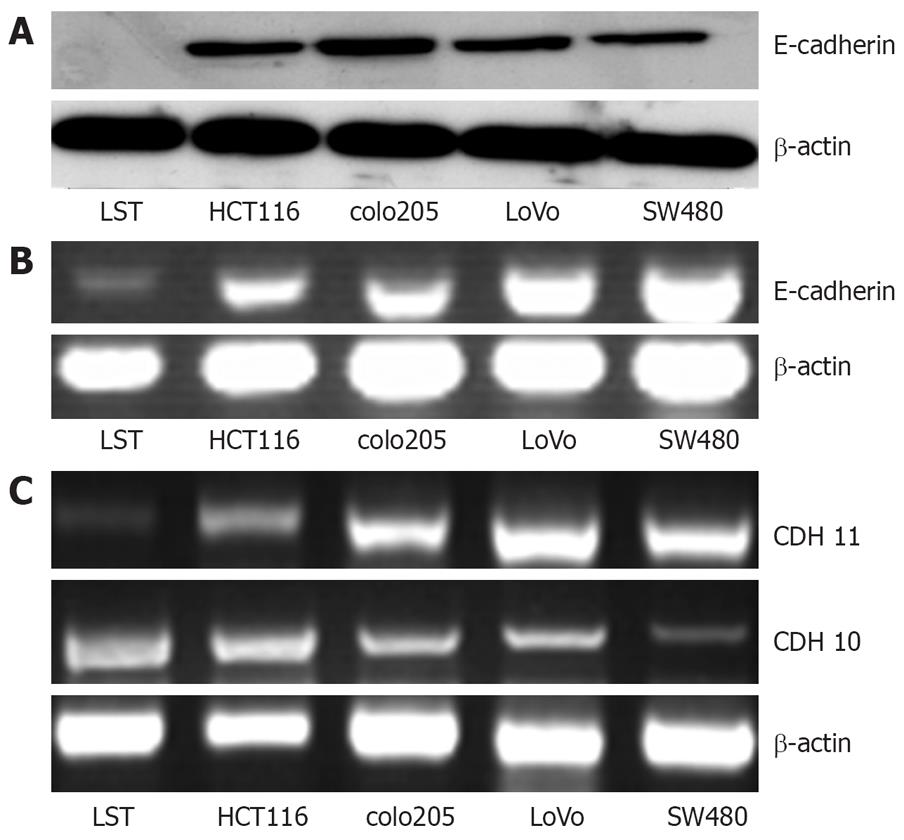
As shown in Tables 2 and 3, the cDNA microarray was applied to analyse the expression patterns of the three cell lines: SW480, lovo, LST. Expression of the cell adhesion molecule E-cadherin in different colon cancer cell lines was confirmed by Western blotting. As shown in Figure 5, the expression of E-cadherin in LST cell line was at a very low level comparing with the other four colon cancer cell lines.
| Accession | Gene name | Gene symbol | Fold change |
| Z24725 | Mitogen inducible 2 | MIG2 | 3.21 |
| Z22555 | CD36 antigen (collagen type I receptor, thrombospondin receptor)-like 1 | CD36L1 | 3.10 |
| S80990 | Ficolin (collagen/fibrinogen domain-containing) 1 | FCN1 | 2.91 |
| AL137480 | Formin binding protein 4 | FNBP4 | 2.73 |
| AL117505 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | ATP2A2 | 2.63 |
| M26683 | Small inducible cytokine A2 (monocyte chemotactic protein 1, homologous to mouse Sig-je) | SCYA2 | 2.45 |
| AF055084 | Monogenic, audiogenic seizure susceptibility 1 homolog (mouse) | MASS1 | 2.39 |
| X68264 | Melanoma cell adhesion molecule | MCAM | 2.38 |
| AF052124 | Secreted phosphoprotein 1 (osteopontin, bone sialoproteinI, early T-lymphocyte activation 1) | SPP1 | 2.31 |
| M25280 | Selectin L (lymphocyte adhesion molecule 1) | SELL | 2.23 |
| M16336 | CD2 antigen (p50), sheep red blood cell receptor | CD2 | 2.19 |
| AL161973 | Myeloid/lymphoid or mixed-lineage leukemia [trithorax (Drosophila) homolog]; translocated to, 4 | MLLT4 | 2.14 |
| L25286 | Collagen, type XV, alpha 1 | COL15A | 2.10 |
| D83018 | Nel (chicken)-like 2 | NELL2 | -48.1 |
| AB040902 | Fibronectin leucine rich transmembrane protein 3 | FLRT3 | -3.51 |
| AA460440 | CD164 antigen, sialomucin | CD164 | -2.23 |
| U85611 | Calcium and integrin binding protein (DNA-dependent protein kinase interacting protein) | CIB1 | -2.03 |
| Accession | Gene name | Gene symbol | Fold change |
| S80990 | Ficolin (collagen/fibrinogen domain-containing) 1 | FCN1 | 42.87 |
| AL161973 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax (Drosophila) homolog); translocated to, 4 | MLLT4 | 26.47 |
| M26683 | Small inducible cytokine A2 (monocyte chemotactic protein 1, homologous to mouse Sig-je) | SCYA2 | 24.92 |
| M31210 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | EDG1 | 11.07 |
| Z22555 | CD36 antigen (collagen type I receptor, thrombospondin receptor)-like 1 | CD36L1 | 4.33 |
| M16336 | CD2 antigen (p50), sheep red blood cell receptor | CD2 | 3.07 |
| U43901 | Laminin receptor 1 (67 kD, ribosomal protein SA) | LAMR1 | 2.55 |
| M85289 | Heparan sulfate proteoglycan 2 (perlecan) | HSPG2 | 2.11 |
| Y13323 | ADAM-like, decysin 1 | ADAMDEC1 | -2.03 |
| AL133035 | Filamin-binding LIM protein-1 | FBLP1 | -2.10 |
| NM_002292 | Laminin, beta 2 (laminin S) | LAMB2 | -2.10 |
| AL161972 | Intercellular adhesion molecule 2 | ICAM2 | -2.12 |
| BE301622 | Pinin, desmosome associated protein | PNN | -2.17 |
| AF053944 | AE-binding protein 1 | AEBP1 | -2.21 |
| Y10931 | Symplekin | SYMPK | -2.24 |
| M98457 | CD209 antigen | CD209 | -2.25 |
| D87469 | Cadherin, EGF LAG seven-pass G-type receptor 2, flamingo (Drosophila) homolog | CELSR2 | -2.26 |
| AW843882 | Leucine-rich repeats and immunoglobulin-like domains 3 | LRIG3 | -2.26 |
| X78947 | Connective tissue growth factor | CTGF | -2.33 |
| AF135021 | Catenin (cadherin-associated protein), alpha-like 1 | CTNNAL1 | -2.36 |
| AL136139 | Enhancer of filamentation 1 (cas-like docking; Crk-associated substrate related) | HEF1 | -2.36 |
| AF055084 | Monogenic, audiogenic seizure susceptibility 1 homolog (mouse) | MASS1 | -2.37 |
| AF039747 | Cadherin 10, type 2 (T2-cadherin) | CDH10 | -2.39 |
| U58516 | Milk fat globule-EGF factor 8 protein | MFGE8 | -2.41 |
| D49742 | Hyaluronan-binding protein 2 | HABP2 | -2.42 |
| U79716 | Reelin | RELN | -2.42 |
| Y11307 | Cysteine-rich, angiogenic inducer, 61 | CYR61 | -2.45 |
| AB007865 | Fibronectin leucine rich transmembrane protein 2 | FLRT2 | -2.48 |
| NM_006149 | Lectin, galactoside-binding, soluble, 4 (galectin 4) | LGALS4 | -2.52 |
| M59807 | Natural killer cell transcript 4 | NK4 | -2.59 |
| AJ009985 | Annexin A9 | ANXA9 | -2.61 |
| NM_014000 | Vinculin | VCL | -2.63 |
| AL117505 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | ATP2A2 | -2.64 |
| AF062075 | Leupaxin | LPXN | -2.67 |
| X05610 | Collagen, type IV, alpha 2 | COL4A2 | -2.70 |
| D12676 | CD36 antigen (collagen type I receptor, thrombospondin receptor)-like 2 (lysosomal integral membrane protein II) | CD36L2 | -2.72 |
| D21255 | Cadherin 11, type 2, OB-cadherin (osteoblast) | CDH11 | -2.77 |
| U70136 | Proteoglycan 4, (megakaryocyte stimulating factor, articular superficial zone protein) | PRG4 | -2.77 |
| S53911 | CD34 antigen | CD34 | -2.82 |
| AW025274 | Kin of IRRE like 2 (Drosophila) | KIRREL2 | -2.85 |
| M73255 | Vascular cell adhesion molecule 1 | VCAM1 | -2.86 |
| L25286 | Collagen, type XV, alpha 1 | COL15A1 | -2.87 |
| AL117604 | Deleted in liver cancer 1 | DLC1 | -2.91 |
| AJ132445 | Claudin 14 | CLDN14 | -2.95 |
| AL133035 | Filamin binding LIM protein 1 | FBLP1 | -3.14 |
| Z24725 | Mitogen inducible 2 | MIG2 | -3.15 |
| X15880 | Collagen, type VI, alpha 1 | COL6A1 | -3.53 |
| AL137480 | Formin binding protein 4 | FNBP4 | -3.58 |
| AF052124 | Secreted phosphoprotein 1 (osteopontin, bone sialoproteinI, early T-lymphocyte activation 1) | SPP1 | -3.60 |
| AB018269 | Calsyntenin 3 | CLSTN3 | -3.70 |
| AB002298 | PDZ domain containing 3 | PDZK3 | -3.97 |
| X87838 | Catenin (cadherin-associated protein), beta 1 (88 kDa) | CTNNB1 | -4.64 |
Recent molecular studies have demonstrated a different histogenesis of flat and depressed tumor in comparison with polypoid adenomas. Flat and depressed tumor has a more malignant tendency and LST is an important subtype of it[24–27]. In the study of disease genesis, cell line and animal model are useful tools to understanding the biological characteristics. However, until now, there is no report on LST cell line and/or animal model. So there is an absolute need to establish in vitro model systems to study the distinctive behavior of LST and the potential mechanism of colon carcinogenesis.
In the present study, we report the characterization of LST-R1 from a fresh laterally spreading tumor specimen. Although the tissue, which the cell line derived from, is villous adenoma accompanied by severe atypical hyperplasia, our data showed it has an obvious malignant tendency. First, LST-R1 cells were passaged more than 110 times without the use of any immortalizing agent; second, the karyotypic analysis showed the cells were triploid with structural chromosome aberrations and a number of numerical abnormalities; third, the tissue from the inoculated nude mice are poorly differentiated adenocarcinoma, which suggests the cell line is originated from malignant clones. The most possible reason for this difference could be that the pathologic type of the tissue (60 mm × 70 mm) was large and uneven, and only a little part of it has been taken for histopathological study while most of the tissue was taken for cell culture. Analysis from only a small number of serial sections might lead to the missed diagnosis of the cancer. Normal cells proliferate at a slower rate than the malignant cells, therefore only the malignant cells left after several passages.
The epithelial and colorectal origin of these cells were confirmed by positive ESA and CK20 staining, miniville and tight junction observed under the electron microscope, as well as morphological observation. After 15 d of inoculation in nude mice, the tumors occurred and grew vertically rather than laterally spreading. The reason we proposed is associated with the different circumstances between hypodermic and colon lumen mucosa. Although the lack of lateral growth in nude mice limits its utility as an in vivo model, it may prove otherwise as an in vitro model system to investigate biological behaviors and malignancy-associated phenotypes of the disease at its onset.
The superficial colon tumor is more likely to invade the mucous membranes and developed cancer[2]. So we compared invasion ability between LST-R1 and other two colorectal cancer cell lines derived from prudent colon cancer with the Matrigel invasion chamber. SW480 is human colorectal adenocarcinoma (Duke’ type B). Lovo is human colorectal adenocarcinoma derived from a fragment of a metastatic tumor nodule in the left supraclavicular region (Duke’ type C, grade IV). Our results demonstrated that the number of LST cells invading through the membrane was more than the other two cell lines and showed that the LST-R1 cells had a high ability of invasion, which was in consistent with the high malignancy of LST.
It is well known that the low expression of the adhesion function contributes to the increasing invasive ability of colon cancer. To explore the reason of higher invasion of LST-R1 cell line, we compared the gene expression profile of LST-R1 cells with SW480 and lovo cells by cDNA microarray. According to cDNA microarray data, a number of genes associated with cell adhesion changed, such as E-cadherin. E-cadherin is a 120-kDa transmembrane glycoprotein that belongs to a large superfamily of calcium-dependent cadherin adhesion molecules[27] and plays an important role in embryogenesis and in mature epithelia. Abnormalities of E-cadherin expression have been described in colorectal carcinoma[28] and have been correlated with increased invasiveness, metastatic potential and poor patient prognosis in this tumor type[29]. Recent research showed that E-cadherin expression was significantly lower in the de novo group tissue than that in the prudent group[3031]. In the present study, by Western blotting analysis, we also confirmed that LST-R1 cells have a prominent lower expression of E-cadherin than that in several other cell lines. Our data therefore suggest that the high invasive ability of LST-R1 cell line has a close relationship with the low expression of adhesion molecule. Further investigations should be taken to study the role of the adhesion molecules (such as E-cadherin) in determining the malignancy of LST.
In conclusion, we established a stable cell line (LST-R1) derived from an early colon cancer-LST and this cell line has an obvious malignant tendency. LST-R1 is also characterized with high invasive ability, and this property may result from the loss of adhesion molecules such as E-cadherin.
Invasion is an important characteristic of malignant tumor and the mechanism of invasion is not very clear. Many genes had been verified to be involved in invasion of cancer including E-cadherin. Cell lines are useful tools for molecular level study and many cell lines have been established for various studies.
Laterally spreading tumor (LST) is a unique subtype of superficial colorectal tumor and in some studies, it has been reported that it had a higher carcinogenesis rate. Invasion is a pivotal step of colorectal cancer carcinogenesis progression and E-cadherin is an important invasion-associated gene. We established a colorectal cell line with high invasion ability and a low expression of E-cadherin.
In this study, we establish a cell line derived from a laterally spreading tumor tissue. Our data suggested that this cell line is a colorectal cell line and has high invasion ability compared with SW480 and Lovo cell lines. There is a loss of E-cadherin expression in Laterally Spreading Tumor-Rectum 1 ( LST-R1) cells. This cell line will be a useful tool for the study of colorectal cancer invasion mechanism.
This cell line is a useful tool for colorectal cancer invasion mechanism study. We will investigate the role of E-cadherin in LST-R1 cells and the relationship between low expression of E-cadherin and high invasion ability.
SKY in this study means metaphase chromosome preparation and spectral karyotyping.
This article is interesting. In this article, the authors characterized a laterally spreading tumor cell line and compare it to several typical colorectal cancer cell lines. Since these tumors progress in a different fashion than typical adenoma to carcinoma colorectal tumors that protrude into the gut lumen, the isolation and comparison of expression of growth patterns of these cells is important.
| 2. | Teixeira CR, Tanaka S, Haruma K, Yoshihara M, Sumii K, Kajiyama G, Shimamoto F. Flat-elevated colorectal neoplasms exhibit a high malignant potential. Oncology. 1996;53:89-93. |
| 3. | Aoki T, Takeda S, Yanagisawa A, Kato Y, Ajioka Y, Watanabe H, Kudo S, Nakamura Y. APC and p53 mutations in de novo colorectal adenocarcinomas. Hum Mutat. 1994;3:342-346. |
| 4. | Ajioka Y, Watanabe H, Kazama S, Hashidate H, Yokoyama J, Yamada S, Takaku H, Nishikura K. Early colorectal cancer with special reference to the superficial nonpolypoid type from a histopathologic point of view. World J Surg. 2000;24:1075-1080. |
| 5. | Hasegawa H, Ueda M, Furukawa K, Watanabe M, Teramoto T, Mukai M, Kitajima M. p53 gene mutations in early colorectal carcinoma. De novo vs. adenoma-carcinoma sequence. Int J Cancer. 1995;64:47-51. |
| 6. | Kudo S, Kashida H, Tamura T. Early colorectal cancer: flat or depressed type. J Gastroenterol Hepatol. 2000;15 Suppl:D66-D70. |
| 7. | Tanaka S, Haruma K, Oka S, Takahashi R, Kunihiro M, Kitadai Y, Yoshihara M, Shimamoto F, Chayama K. Clinicopathologic features and endoscopic treatment of superficially spreading colorectal neoplasms larger than 20 mm. Gastrointest Endosc. 2001;54:62-66. |
| 8. | Okamoto T, Tanaka S, Haruma K, Hiraga Y, Kunihiro M, Goishi H, Tanimoto T, Sumii M, Yoshihara M, Sumii K. Clinicopathologic evaluation on colorectal laterally spreading tumor (LST). Nippon Shokakibyo Gakkai Zasshi. 1996;93:83-89. |
| 9. | Kudo S. Endoscopic mucosal resection of flat and depressed types of early colorectal cancer. Endoscopy. 1993;25:455-461. |
| 10. | Kusaka T, Fukui H, Sano Y, Ueda Y, Chiba T, Fujimori T. Analysis of K-ras codon 12 mutations and p53 overexpression in colorectal nodule-aggregating tumors. J Gastroenterol Hepatol. 2000;15:1151-1157. |
| 11. | Strekalovsky VP. Results of endoscopic removal of villous tumors of the colon. Endoscopy. 1983;15:49-52. |
| 12. | Yasuda K, Ajioka Y, Watanabe H, Matsuda K, Kitano S. Morphogenesis and development of superficial spreading tumor of the colon and rectum. Pathol Int. 1997;47:769-774. |
| 13. | Rubesin SE, Saul SH, Laufer I. Carpet lesions of the colon. Radiographics. 1985;5:537-552. |
| 14. | Kudo S, Kashida H, Nakajima T, Tamura S, Nakajo K. Endoscopic diagnosis and treatment of early colorectal cancer. World J Surg. 1997;21:694-701. |
| 15. | Ohno Y, Terai T, Ogihara T, Hirai S, Miwa H. Laterally spreading tumor: clinicopathological study in comparison with the depressed type of colorectal tumor. J Gastroenterol Hepatol. 2001;16:770-776. |
| 16. | Teixeira CR, Tanaka S, Haruma K, Yoshihara M, Sumii K, Kajiyama G, Shimamoto F. Flat-elevated colorectal neoplasms exhibit a high malignant potential. Oncology. 1996;53:89-93. |
| 17. | Jiang B, Liu SD, Zhi FC, Pan DS, Zhou D, Wan TM, Zhou DY. The diagnosis and treatment of 25 cases of laterally spreading tumor of the large intestine. Diyi Junyi Daxue Xuebao. 2002;22:189-191. |
| 18. | Kudo S. Early Colorectal Cancer-Detection of Depressed Types of Colorectal Carcinoma. Igaku-Shoin Medical Publisher. 1996;50-51. |
| 19. | Deng W, Tsao SW, Lucas JN, Leung CS, Cheung AL. A new method for improving metaphase chromosome spreading. Cytometry A. 2003;51:46-51. |
| 20. | An International System for Human Cytogenetic Nomenclature (1985) ISCN 1985. Report of the Standing Committee on Human Cytogenetic Nomenclature. Birth Defects Orig Artic Ser. 1985;21(1):1-117. |
| 21. | Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47:3239-3245. |
| 22. | Wang XY, Jiang B, Lai ZS, Ma WM, Geng Y. Screening of differentially expressed genes related to laterally spreading tumor by cDNA microarray. Diyi Junyi Daxue Xuebao. 2004;24:1023-1025. |
| 23. | Wang X, Li M, Wang J, Yeung CM, Zhang H, Kung HF, Jiang B, Lin MC. The BH3-only protein, PUMA, is involved in oxaliplatin-induced apoptosis in colon cancer cells. Biochem Pharmacol. 2006;71:1540-1550. |
| 24. | Watanabe T, Muto T. Colorectal carcinogenesis based on molecular biology of early colorectal cancer, with special reference to nonpolypoid (superficial) lesions. World J Surg. 2000;24:1091-1097. |
| 25. | Fujimori T, Satonaka K, Yamamura-Idei Y, Nagasako K, Maeda S. Non-involvement of ras mutations in flat colorectal adenomas and carcinomas. Int J Cancer. 1994;57:51-55. |
| 26. | Yamagata S, Muto T, Uchida Y, Masaki T, Sawada T, Tsuno N, Hirooka T. Lower incidence of K-ras codon 12 mutation in flat colorectal adenomas than in polypoid adenomas. Jpn J Cancer Res. 1994;85:147-151. |
| 27. | Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551-572. |
| 28. | El-Bahrawy MA, Pignatelli M. E-cadherin and catenins: molecules with versatile roles in normal and neoplastic epithelial cell biology. Microsc Res Tech. 1998;43:224-232. |
| 29. | Van Aken J, Cuvelier CA, De Wever N, Roels J, Gao Y, Mareel MM. Immunohistochemical analysis of E-cadherin expression in human colorectal tumours. Pathol Res Pract. 1993;189:975-978. |
| 30. | Streit M, Schmidt R, Hilgenfeld RU, Thiel E, Kreuser ED. Adhesion receptors in malignant transformation and dissemination of gastrointestinal tumors. J Mol Med. 1996;74:253-268. |
| 31. | Wlodarczyk J, Bethke B, Mueller E, Stolte M, Mueller J. A comparative study of E-cadherin and stromelysin-3 expression in de novo and ex adenoma carcinoma of the colorectum. Virchows Arch. 2001;439:756-761. |