Published online Mar 14, 2008. doi: 10.3748/wjg.14.1588
Revised: December 6, 2007
Published online: March 14, 2008
AIM: To comprehensively identify the proteins of tumor relative antigen Ca-Hb3 recognized by colorectal carcinoma monoclonal antibody Hb3.
METHODS: Ca-Hb3 was isolated by SDS-polyacrylamide gel electrophoresis (PAGE) followed by digestion with trypsin. Trypsin peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The proteins identified by mass spectrometry were analyzed using bioinformatics.
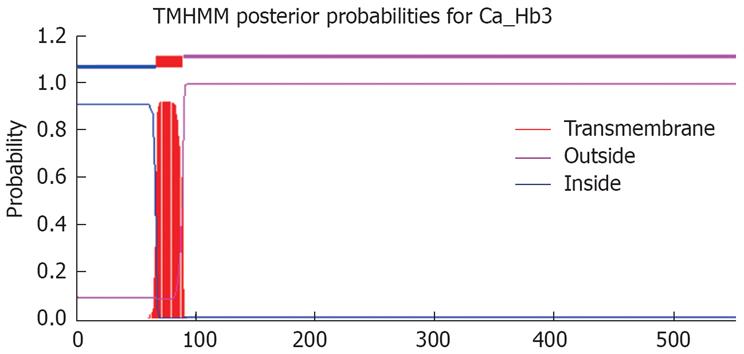
RESULTS: Ca-Hb3 was identified as a CKAP4-like protein by Nano HPLC tandem mass spectrometry analysis. The molecular weight of CKAP4-like protein was 62.02 kDa, including one hydrophobic region, one transmembrane domain, five coiled coils, four glycosylation sites and forty-nine phosphorylation sites. CKAP4-like protein had a high homogeneity with DeltaNp63α. The characteristic expression of DeltaNp63α that is considered a potential oncogene in the isoforms of p63 was similar to that of Ca-Hb3.
CONCLUSION: Ca-Hb3 is probably a CKAP4-like protein, belonging to DeltaNp63α isoform of p63 family.
- Citation: Sun S, Guo FJ, Tong YQ, Zhu JG, Li GC. Colorectal carcinoma-associated antigen Ca-Hb3 detected by one-dimensional SDS-polyacrylamide gel electrophoresis and liquid chromatography-tandem mass spectrometry. World J Gastroenterol 2008; 14(10): 1588-1591
- URL: https://www.wjgnet.com/1007-9327/full/v14/i10/1588.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.1588
Colorectal cancer can be considered a complex disease with predisposing genetic variants and environmental factors that contribute to the illness as a whole. In recent years, many studies reported that the incidence of colorectal cancer is increased by 4% per year in China[1–3]. Hb3 is an anti-colorectal cancer monoclonal antibody produced in our laboratory. Its sensitivity and specificity are superior to those of anti-CEA. Monoclonal antibody Hb3 (mAb Hb3) targets the Hb3 antigen which is expressed in 85% of colorectal cancers, showing that CA-Hb3 may be useful in the diagnosis of colorectal cancer. In this study, using SDS-polyacrylamide gel electrophoresis (PAGE) and microcapillary reversed-phase liquid chromatography-tandem mass spectrometry, we have identified 5 proteins, including one membrane protein which was initially determined to be Ca-Hb3 by bioinformatics analysis.
mAb Hb3, a murine IgM-type mAb, was prepared using 5 × 108 Hb3 hybridoma cells (our laboratory) to immunize Balb/c mice. Mice were sacrificed and ascites was extracted to obtain supernatant by centrifugation.
NanoLC-MS was performed online with a Waters capillary LC system interfaced with a Waters (Micromass) electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) mass spectrometer.
Human colon carcinoma HRT-18 cells were cultured in RPMI 1640 medium (Promega Inc.) supplemented with 100 mL/L heat-inactivated fetal bovine serum (FBS, Hangzhou Sijiqing BRL Inc.), 30 g/L L-glutamine, 100 kg/L streptomycin and 100 U/L penicillin at 37°C in a humidified atmosphere containing 50 mL/L CO2.
Total cell extraction from human colon carcinoma HRT-18 cells was performed by Western blot assay. Samples in the cold lysate buffer (50 mmol/L Tris-Cl pH 8.0, 150 mmol/L NaCl, 10 mmol/L Triton X-100, 10 mmol/L PMSF) were electrophoresed on 10% gradient SDS-polyacrylamide gels in the presence of β-mercaptoethanol. Half of the proteins were transblotted onto a NC membrane, which was blocked with 50 mg/mL fat-free milk and 100 mL/TBS for 2 h and incubated with primary antibody (mAb Hb3) overnight at 4°C. The membrane was washed and incubated with HRP-conjugated goat anti-mouse IgG/IgM (SIGMA Inc.) at room temperature for 1 h. The membrane was washed and the antigen-antibody reaction was visualized using an ECL detection system (KPL Inc.). The other half of proteins were stained with Coomassie blue.
Bands of interest were excised and minced into 1 mm3 pieces with sterile razor blade. The pieces were washed 3 times with ddH2O and then with 50% acetonitrile in 100 mmol/L NH4HCO3, incubated in water bath for 60 min if necessary. The liquid was discarded, 100 &mgr;L 1% acetonitrile was added and the dried acetonitrile was removed after 10 min. For SDS-PAGE gel bands, gel pieces were rehydrated and proteins were reduced with 200 &mgr;L of 100 mmol/L DTT in 100 mmol/L ammonium bicarbonate for 30 min at 56°C. The reduction solution was discarded and 100 &mgr;L of 50 mmol/L iodoacetamide in 100 mmol/L ammonium bicarbonate was added. Incubation lasted for 30 min at room temperature in the dark. Gel bits were washed with 500 &mgr;L of 25 mmol/L ammonium bicarbonate in 500 mL/L acetonitrile for 15 min, then 100 &mgr;L 100% acetonitrile was added and the dried acetonitrile was removed after 10 min. The gel pieces were rehydrated by adding 5 &mgr;L of trypsin in 40 mmol/L ammonium bicarbonate and incubated overnight at 37°C. The supernatant was removed and placed into a new tube. Peptides were extracted from the gel pieces with 50 &mgr;L of 20 mmol/L ammonium bicarbonate for 20 min and then with 50 &mgr;L of 5% formic acid in 50% acetonitrile for 2 × 20 min. The extracts were added to the supernatant. The mixed solution was dried using a Speed-Vas.
RP-HPLC separation of the peptide samples was performed using a bioinert ultimate nano-HPLC system (Dionex, Sunnyvale, CA, USA). Six microliters of each sample was injected and peptides were purified and concentrated on a C18-PepMap precolumn (0.3 mm ID× 65 mm, 100Å pore size, 3 mm particle size, Dionex) at a flow rate of 0.02 mL/min. Subsequently, peptides were separated on an analytical 75 mm ID × 150 mm C18-PepMap column (Dionex, 100Å pore size, 3 mm particle size) at a column flow rate of 300 mL/min. The ACN gradient (solution A: 1 mL/L formic acid, 20 mL/L ACN; solution B: 1 mL/L formic acid, 800 mL/L ACN) started at 50 mL/L B and ended at 70 mL/L B in 45 min. MS and MS/MS data were collectd using a Micromass Q-TOF micro mass spectrometer (Waters, Milford, MA, USA). Doubly and triply charged peptide ions were automatically chosen by the MassLynx software (Waters) and fragmented for a maximum of 7 s for each component. MS data were automatically processed and peak lists for database search were generated by the MassLynx software. Database search was carried out with an in-house MASCOT server using an IPI protein database (Mouse, version 3.07), for a mass tolerance of 5 nkat and allowance for up to 16.67 nKat trypsin miscleavage and variable amino acid modifications consisting of methionine oxidation and cysteine carbamidomethylation.
Tandem mass spectrometry (MS/MS) data were collected by the Mass-Lynx 4.0 software (Waters), deisotoped and converted by Waters ProteinLynx Global server 1.0 software to “peak list” (pkl) files, which include the mass values, the intensity (at least 5 counts/s) and the charge of precursor ions. The parameters used in the creation of pkl files were the following background subtraction below curve 10%, which was smoothed three times with “smooth window” (channels) 2.0 in the Savitzky Golay mode, centroid at 80% top and minimum peak width at half height at 4. The pkl files were analyzed using a licensed copy of the Mascot2.0 Program (MatrixScience Ltd., London) on a 2.6 GHz Pentium-4 personal computer with 2 GB of RAM, which compared the files against a human protein non-redundant database containing 146724 protein sequences downloaded as FASTA formatted sequences from NCBI. Search parameters were set as follows: enzyme, trypsin, allowance of up to one missed cleavage peptide, mass tolerance, 0.3u MS/MS mass tolerance, fixed modification parameter, carbamoylmethylation (C), variable modification parameter, oxidation (at Met), auto hits allowed (only significant hit report), result format as peptide summary report. Predictions for putative transmembrane domains (TMDs) in all identified proteins were carried out using the transmembrane hidden Markov model (TMHMM) algorithm, available at http://www.cbs.dtu.dk/services/TMHMM.
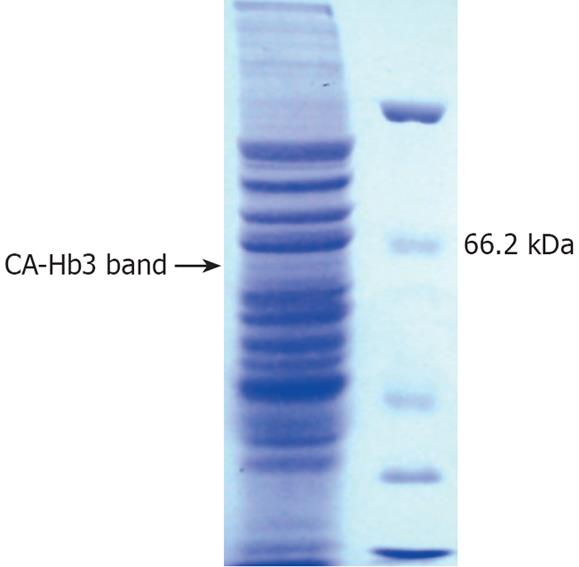
Ca-Hb3 was characterized by Western blot analysis (Figure 1) and one specific electrophoretic band of about 65 kDa in the cell lysates was shown, supporting the previous experiment results[3]. Ca-Hb3 crude antigen was separated by one-dimensional SDS-PAGE (Figure 2). Proteins present in the interest band were digested by trypsin. The tryptic peptide was extracted from the purpose band and further separated by reversed-phase nanoLC, detected and sequenced with a Waters Q-TOF micro mass spectrometer. Raw data were analyzed by Mascot ms/ms ion search against the human nonredundant database. Using the stringent criteria mentioned above, we identified 5 non-redundant proteins: one inquinate hybrid protein, three nuclear proteins and one CKAP4-like protein. The molecular weight of the second protein (Chain A, crystal structure of the Tpr2a domain of Hop in complex with the Hsp90 peptide Meevd) showed 15484u by MS, and was thus judged to be the inquinate hybrid protein. We found that only CKAP4-like protein (similar to cytoskeleton-associated protein 4) belonged to an integral membrane protein (http://www.expasy.org), consistent with the previous experiment results (Figure 3).
Hb3 is an IgM monoclonal antibody against CA-Hb3[1–3]. The results of immunohistochemistry showed that the positive rate of Hb3 reacting with colorectal carcinoma was high.
To characterize the proteins of complex proteomes, the proteins were analyzed by 2D-PAGE followed by MS or MS/MS, or by 2D-LC-MS/MS. Because Ca-Hb3 is located in membrane, it is difficult to resolve membrane proteins by 2D-PAGE. We replaced the 2D-PAGE and 2D-LC with a one-dimensional SDS-PAGE. By applying the method mentioned above, we identified 5 non-redundant proteins. Except for an inquinate hybrid protein and three nuclear proteins, the remaining CKAP4-like protein had a transmembrane domain at the corresponding Ca-Hb3 location.
The p63 gene, independently cloned by multiple laboratories, is a member of the p53 famliy of transcription factors[4–6]. p53 is a tumor suppressor that is inactivated in a majority of human cancers. In response to cellular stresses, such as DNA damage and oncogene activation, p53 is stabilized and acts as a sequence-specific transcription factor, trans-activating target genes are involved in the processes of cell cycle arrest, DNA repair, and apoptosis[7–10]. These proteins exhibit a high sequence and structural similarity to p53, while revealing a considerable functional divergence[11–15].
The human p63 gene consisting of 15 exons, resides on chromosome 3q27-29 and can be expressed in two different promoters[1617]. Transcription from the first and second promoters gives rise to TA or ΔN amino terminus of p63. Both TA and ΔN transcripts can be alternatively split at the carboxy-terminus, leading to formation of α, β and γ isoforms of TA and ΔN p63α. A number of studies have highlighted the oncogenic potential of ΔN p63α which is over-expressed in several epithelial cancers, often as a result of gene amplification[18–23]. Over-expression of ΔN p63α isoform in rat-1A cells increases colony growth in soft agar and xenograft tumor formation in nude mice, supporting that p63 acts as an oncogene[19]. Furthermore, ΔNp63α can prevent p53-mediated trans-activation, growth arrest, and apoptosis[24]. The expression of ΔNp63α is associated with epithelial homeostasis, poor survival of ovarian cancer patients, and promotes keratinocyte adhesion, inhibits apoptosis, and maintains the integrity of epidermal tissue[2526]. ΔNp63α regulates CD44 and keratins 4, 6, 14 and 19 in squamous cell carcinoma of the head and neck, and down-regulation of DeltaNp63α acquires invasive phenotype of human squamous cell carcinoma[2728]. It is hypothesized that ΔNp63α prolongs survival and maintains the proliferating capacity of epithelial stem and cancer cells.
In conclusion, colon cancer monoclonal antibody Hb3 is probably a CKAP4-like protein, belonging to DeltaNp63α isoform of p63 family, which can be considered a marker for colon cancer progression or as a therapeutic target.
Hb3 is an anti-colorectal cancer monoclonal antibody produced in our laboratory. Its sensitivity and specificity are superior to those of anti-carcinoembryonic antigen (anti-CEA), showing that CA-Hb3 might be useful in the diagnosis of colorectal cancer.
There are a number of researchers who are searching for a useful marker of tumor. Antigen, as an effect target, is applied in clinical therapy for colon cancer.
ΔNp63α obtained by one-dimensional polyacrylamide gel electrophoresis (PAGE) and liquid chromatography-tandem mass spectrometry could be considered a marker for colon cancer progression or as a therapeutic target.
ΔNp63α could be applied in treatment of cancer patients as an effect target.
DeltaNp63α is an isoform of P63 family. ΔNp63α is over-expressed in several epithelial cancers, often as a result of gene amplification. Over-expression of a ΔNp63α isoform in rat-1A cells increases colony growth in soft agar and xenograft tumor formation in nude mice.
This paper is well organized with valuable conclusions.
| 1. | Sun QB, Ho JJ, Kim YS. Human colonic cancer associated antigens detected by three monoclonal antibodies. Chin Med J (Engl). 1986;99:63-74. |
| 2. | Cheng ZC, Zhou ZJ, Jiang YX, Sun QB. Preliminary study on differentiating colon carcinoma from adenoma by monoclonal antibody Hb3. Zhonghua Zhongliu Zazhi. 1988;10:29-31. |
| 3. | Hu JY, Su JZ, Pi ZM, Zhu JG, Zhou GH, Sun QB. Radioimmunoimaging of colorectal cancer using (99m)Tc labeled monoclonal antibody. World J Gastroenterol. 1998;4:303-306. |
| 4. | Augustin M, Bamberger C, Paul D, Schmale H. Cloning and chromosomal mapping of the human p53-related KET gene to chromosome 3q27 and its murine homolog Ket to mouse chromosome 16. Mamm Genome. 1998;9:899-902. |
| 5. | Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305-316. |
| 6. | Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839-843. |
| 8. | Stewart ZA, Pietenpol JA. p53 Signaling and cell cycle checkpoints. Chem Res Toxicol. 2001;14:243-263. |
| 9. | Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303-312. |
| 10. | Li YN, He ZX, Liu LK, He HW. Function of P63 on the development of salivary glands. Huaxi Kouqiang Yixue Zazhi. 2007;25:111-114. |
| 11. | Bourdon JC. p53 and its isoforms in cancer. Br J Cancer. 2007;97:277-282. |
| 12. | Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165-168. |
| 13. | Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962-972. |
| 14. | Marabese M, Marchini S, Marrazzo E, Mariani P, Cattaneo D, Fossati R, Compagnoni A, Signorelli M, Moll UM, Codegoni AM. Expression levels of p53 and p73 isoforms in stage I and stage III ovarian cancer. Eur J Cancer. 2008;44:131-141. |
| 15. | Nedelcu AM, Tan C. Early diversification and complex evolutionary history of the p53 tumor suppressor gene family. Dev Genes Evol. 2007;217:801-806. |
| 16. | Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1:199-207. |
| 17. | Hu H, Xia SH, Li AD, Xu X, Cai Y, Han YL, Wei F, Chen BS, Huang XP, Han YS. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer. 2002;102:580-583. |
| 18. | Mills AA. p63: oncogene or tumor suppressor? Curr Opin Genet Dev. 2006;16:38-44. |
| 20. | Malaguarnera R, Mandarino A, Mazzon E, Vella V, Gangemi P, Vancheri C, Vigneri P, Aloisi A, Vigneri R, Frasca F. The p53-homologue p63 may promote thyroid cancer progression. Endocr Relat Cancer. 2005;12:953-971. |
| 21. | Choi HR, Batsakis JG, Zhan F, Sturgis E, Luna MA, El-Naggar AK. Differential expression of p53 gene family members p63 and p73 in head and neck squamous tumorigenesis. Hum Pathol. 2002;33:158-164. |
| 22. | Park BJ, Lee SJ, Kim JI, Lee SJ, Lee CH, Chang SG, Park JH, Chi SG. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 2000;60:3370-3374. |
| 23. | Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1:199-207. |
| 24. | Westfall MD, Pietenpol JA. p63: Molecular complexity in development and cancer. Carcinogenesis. 2004;25:857-864. |
| 25. | Lee HO, Lee JH, Kim TY, Lee H. Regulation of DeltaNp63alpha by tumor necrosis factor-alpha in epithelial homeostasis. FEBS J. 274(24):6511-6522. |
| 26. | Voroteliak EA, Chermnykh ES, Tkachenko SB, Vasil'ev AV, Terskikh VV. Expression and function of p63 gene in epithelial cells. Izv Akad Nauk Ser Biol. (4):389-393. |
| 27. | Boldrup L, Coates PJ, Gu X, Nylander K. DeltaNp63 isoforms regulate CD44 and keratins 4, 6, 14 and 19 in squamous cell carcinoma of head and neck. J Pathol. 2007;213:384-391. |
| 28. | Higashikawa K, Yoneda S, Tobiume K, Taki M, Shigeishi H, Kamata N. Snail-induced down-regulation of DeltaNp63alpha acquires invasive phenotype of human squamous cell carcinoma. Cancer Res. 2007;67:9207-9213. |