Published online Feb 28, 2007. doi: 10.3748/wjg.v13.i8.1195
Revised: October 25, 2006
Accepted: December 9, 2006
Published online: February 28, 2007
AIM: To evaluate the implication of substitutions in the hepatitis C virus (HCV) non-structural 5A (NS5A) protein in the resistance of HCV during mono-interferon (IFN) or combined IFN-ribavirin (IFN-R) therapy. Although NS5A has been reported to interact with the HCV RNA-dependent RNA polymerase, NS5B, as well as with many cellular proteins, the function of NS5A in the life cycle of HCV remains unclear.
METHODS: HCV quasispecies were studied by cloning and sequencing of sequential isolates from patients infected by HCV genotype 1b. Patients were treated by IFN-α2b for 3 mo followed by IFN-α2b alone or combined IFN-R therapy for 9 additional months. Patients were categorized intro two groups based on their response to the treatments: 7 with sustained virological response (SVR) (quasispecies = 150) and 3 non-responders (NR) to IFN-R (quasispecies = 106).
RESULTS: Prior to treatment, SVR patients displayed a lower complexity of quasispecies than NR patients. Most patients had a decrease in the complexity of quasispecies during therapy. Analysis of amino acids substitutions showed that the degree of the complexity of the interferon sensitivity-determining region (ISDR) and the V3 domain of NS5A protein was able to discriminate the two groups of patients. Moreover, SVR patients displayed more variability in the NS5A region than NR patients.
CONCLUSION: These results suggest that detailed molecular analysis of the NS5A region may be important for understanding its function in IFN response during HCV 1b infection.
- Citation: Veillon P, Payan C, Le Guillou-Guillemette H, Gaudy C, Lunel F. Quasispecies evolution in NS5A region of hepatitis C virus genotype 1b during interferon or combined interferon-ribavirin therapy. World J Gastroenterol 2007; 13(8): 1195-1203
- URL: https://www.wjgnet.com/1007-9327/full/v13/i8/1195.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i8.1195
In Western countries, the hepatitis C virus (HCV) is the major cause of chronic hepatitis, cirrhosis and hepatocellular carcinoma[1]. HCV is a RNA virus belonging to the Flaviviridae. It has a single-stranded plus-sense genome of approximately 9.6 kb with a single open reading frame (ORF) encoding four structural (C, E1, E2 and p7) and six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B). Current combination therapy of pegylated interferon (IFN) alpha and ribavirin (IFN-R) is not universally effective in patients with chronic hepatitis C; patients infected with genotypes 2 or 3 show a high rate of sustained virological response (SVR) (90%) whereas the HCV genotype 1 infected patients have the lowest level of SVR (40% to 50%)[2-4]. Factors that can predict the response to antiviral therapy are not well established; the accepted predictive parameters are: age, sex, pre-treatment viral load, fibrosis stage and HCV genotype[5]. Numerous studies have been undertaken to explain the resistance of genotype 1-infected patients to IFN therapy. Causes and mechanisms are not well understood, but several viral genomic regions have been suggested to antagonize the antiviral effect of IFN alpha, such as the E2, NS3/4A and NS5A regions[6].
The non-structural 5A (NS5A) protein is the ninth protein coded by the ORF of HCV. It has a length of 447 amino acids (aa) for genotype 1b. An interaction with the double-stranded RNA protein kinase (PKR) has been described for NS5A within the codons 2209 and 2274 (NS5A2209-2274)[7-9]. This interaction can block the IFN signalling pathway in cultured cells, and certain authors have suggested that this mechanism may be implicated in HCV resistance to IFN therapy. In 1995, Enomoto et al[10] described a correlation between the number of mutations within a 40 aa sequence of the NS5A region and the response to IFN therapy in genotype 1b-infected patients. This sequence has been termed the interferon sensitivity-determining region (ISDR). These results were then confirmed by other Japanese studies[11-15], but were never in accordance with most Western European[16-20] and American studies[21-23], in which most of the ISDR sequences harbored an intermediate profile. A recent meta-analysis focusing on the number of mutations within NS5A ISDR confirmed the predictive usefulness of ISDR, but a real geographical difference between Caucasian and Asiatic patients was underlined[24].
Recently, it was suggested that NS5A protein may also inhibit the antiviral effect of IFN in a PKR-independent manner in a human hepatocytic cell line[25]. Furthermore, another domain in the NS5A region, named V3 (NS5A2356-2379)[26], could also be linked to IFN resistance. It has been suggested that mutations in the V3 domain may correlate with IFN response[17]. However, two reports have shown that insertion and deletion around the ISDR and the V3 domain had no impact on HCV RNA replication in cell culture[27,28].
In the present study, we examined mutations in the complete NS5A region, including its ISDR, PKR binding domain (PKRbd) and V3 domain, of HCV genotype 1b from patients treated with IFN or combined IFN-R therapy. HCV quasispecies of ten patients were selected for the analysis based on their response to the treatments combined IFN-R therapy.
We analyzed PCR products from ten HCV genotype 1b infected patients included in a therapeutic trial[29]. Seven of them had SVR - three early and four slow responders according to HCV RNA clearance during therapy at mo 3 (M3) - and three were non-responders to IFN-R (NR). Patients were all studied at baseline (D0), M3 and M6, when gene amplification was possible (positive PCR). Patients received 6 million units of IFN-α2b three times weekly. If HCV RNA was still detectable at M2, ribavirin was combined with IFN at M3 and continued up to M12. SVR was defined as undetectable HCV RNA during treatment and six months after the end of therapy (EOT). Non-SVR was defined as detectable HCV RNA during treatment and six months after the EOT.
Quantification of serum HCV RNA was performed using VERSANT HCV RNA 3.0 assay (Bayer Diagnostics, Emeryville, CA, USA) with a detection threshold at 615 IU/mL (2.79 log IU/mL).
HCV RNA was extracted from 200 μL of serum by guanidinium thiocyanate and silica method[30]. The full-length NS5A gene was amplified by nested RT-PCR. RT was combined with the first round of PCR with outer primers E1 and E2 while second amplification was performed with inner primers I3 and I4 as previously described (Table 1)[17,31]. PCR products were then subjected to electrophoresis in a 1% agarose gel (NuSieve GTG, FMC) and visualized by ethidium bromide staining before purification. In this study, we sequenced 343 296 nts from 256 clones of the complete NS5A region. According to Taq error rate (0.182 × 10-4)[32], 6.247 RT-PCR-introduced errors could have been analyzed as substitutions for all clones, i.e. 0.024 RT-PCR-introduced errors per clone.
| Forward primers | Positions nt | |
| E1 | 5' GAGGGGGCTGTGCAGTGGATG 3' | 6057-6077 |
| I3 | 5' TCCGGCTCGTGGCTAAGGGA 3' | 6246-6265 |
| S2 | 5' GGATTTCCACTACGTGACGGG 3' | 6620-6640 |
| S4 | 5' GGGTCTCCCCCCTCCTTGGCC 3' | 6906-6926 |
| S31 | 5' GGACTACGTCCCTCCGGTGG 3' | 7241-7260 |
| Reverse primers | ||
| AS2 | 5' CGTCACGTAGTGGAAATCCCC 3' | 6618-6638 |
| AS32 | 5' GGGACGTAGTCCGGGTCCTTC 3' | 7232-7252 |
| I4 | 5' GCAGCAGACGAGATCCTCAC 3' | 7567-7586 |
| E2 | 5' GCTGCGAGATGTTGTGGCGTA 3' | 7698-7718 |
| Cloning primers | (pMOS vector) | |
| SeqS1 | 5' GGGAAAGCTTGCATGCCTGC 3' | |
| SeqAS2 | 5' GACGTTGTAAAACGACGGCC 3' |
PCR fragments cut out from the agarose gel were purified using a mini-column system (Wizard PCR Preps DNA purification system, Promega) and then were ligated into 50 ng of pMOS vector (pMOSBlue blunt-ended cloning kit, Amersham Bioscience). Transformants were grown on LB-agar plates containing 100 μg/mL ampicillin and 15 μg/mL tetracycline. The presence of HCV insert was confirmed by plasmid amplification using SeqS1 and SeqAS2 primers (Table 1). We produced between 4 and 17 clones per patient.
Cycle sequencing was performed using the CEQ 8000 Dye Terminator Cycle Sequencing kit following the manufacturer’s instructions (Beckman Coulter) before automatic electrophoresis of the sequencing products using a CEQ 8000 (Beckman Coulter). Inner PCR primers S2, S4, S31, AS2 and AS32 were used as sequencing primers (Table 1). Sequences were analyzed using the CEQ 8000 software.
Multiple nucleotide (nt) sequence alignment and ambiguity were carried out with CLUSTAL X interface[33]. The nucleotide substitution rate over sites within each set of variants was estimated by using DAMBE software. The variability at each site i was measured by calculating the Shannon entropy at the site i (Hi) with the following formula: Hi = -(∑4j=1 pj log2 pj), where j = 1, 2, 3, 4 corresponding to nucleotide A, C, G and T, and pj is the proportion of nucleotide j at site i. The normalized entropy Sn was calculated as Sn = H/log N, where N is the total number of analysed sequences[34]. Types of mutational changes were also determined by means of MEGA 2.3 software. Genetic distances between pairs of sequences were performed using Kimura 2-parameters method[35]. The frequency of synonymous (dS) and non-synonymous (dN) substitutions per site were calculated with Nei-Gojobori method using the Jukes-Cantor correction to account for multiple substitution at the same site[36]. The dN/dS ratio is a measure of immunity pressure on a region. Construction of the phylogenetic tree of NS5A variants obtained from ten patients was performed using MEGA 2.3 software. The phylogenetic tree was constructed with the neighbour-joining method and bootstrap resampling (1000 replicates were used to test reliability of the tree topology)[37]. Master sequence was defined as the sequence obtained from direct sequencing of PCR products obtained before cloning.
A multiple alignment of the translated aa sequences was generated. Alignment analysis of nucleotides and proteins was performed using HCV-J (genotype 1b, D90208) as reference and HCV-H (genotype 1a, M67463) as out-group for the construction of the phylogenetic tree.
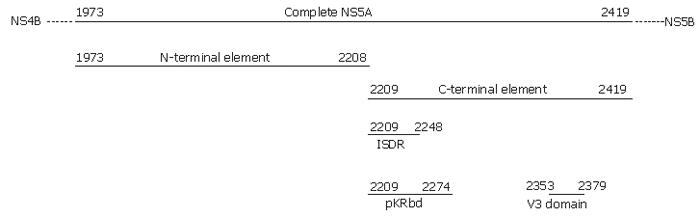
Six regions were studied: the ISDR (NS5A2209-2248), PKR binding domain (PKRbd) (NS5A2209-2274), V3 (NS5A2353-2379), the N-terminal element of NS5A protein until ISDR (NS5A1973-2208), the C-terminal element of NS5A protein from ISDR to the end (NS5A2209-2419), and the complete sequence of NS5A (NS5A1973-2419) (Figure 1).
Values for quantitative variables were expressed as means ± SD. Comparisons between groups were performed using the Student T-test or the non-parametric Mann-Whitney U-test or Kruskal-Wallis K-test for quantitative variables. The Pearson’s correlation coefficient was used. A P-value of less than 0.05 was considered to be statistically significant.
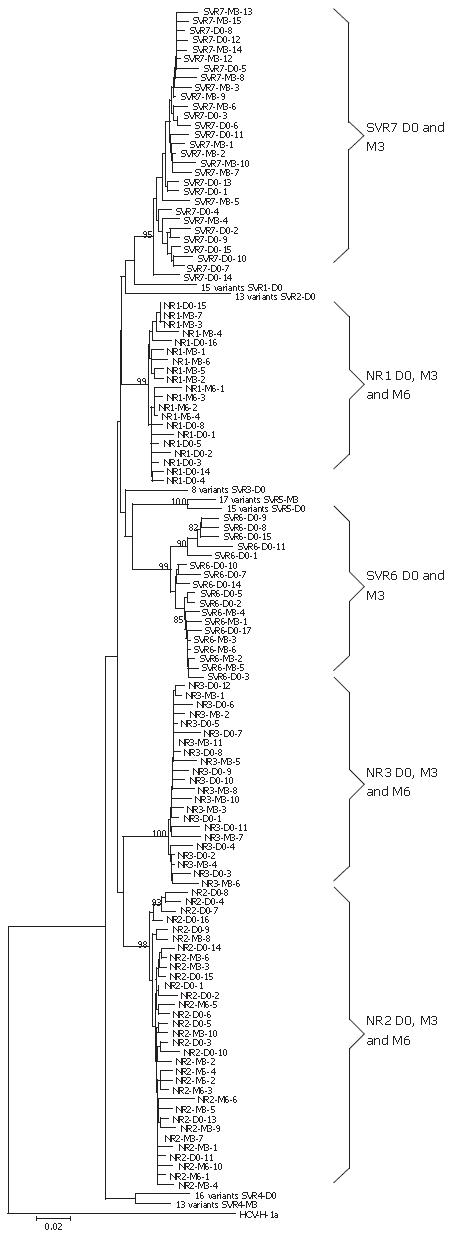
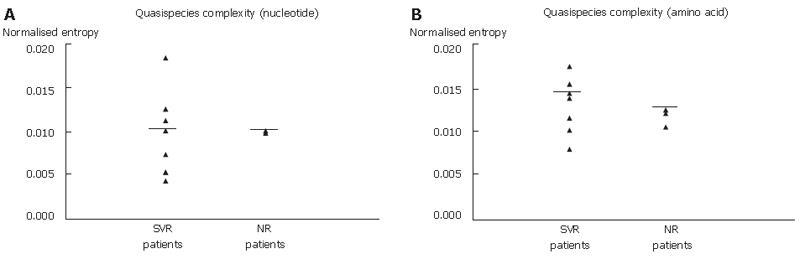
Characteristics of each patient are summarized in Table 2. We studied ten patients (four males and six females). Average patient age was 49.5 years and HCV viral load was 5.35 log IU/mL (mean at baseline). We found a significant difference in viral load between the three groups of early, slow SVR and NR (P = 0.034), whereas no significant difference was found between the SVR and NR groups at baseline. The genetic variability of the NS5A region was confirmed by cloning NS5A PCR products obtained from the ten patients (7 SVR and 3 NR) into the pMOS vector. In total, PCR products from 256 clones of the NS5A region were analyzed (Table 2). No cluster in correlation with treatment response was observed in the phylogenetic tree (Figure 2). However, in two slow SVR patients (SVR4 and SVR5) two different clusters were individualized with two populations of variants (D0 and M3, respectively). In the two other slow SVR patients (SVR6 and SVR7) and in NR patients, we found mixed populations of variants at the beginning of treatment (D0) and during treatment (M3 or M6). Genetic complexity of nt and aa sequences calculated by Shannon entropy showed no significant difference between the two groups studied (Figure 3, A and B). Interestingly, when we looked at early SVR in comparison with slow SVR, we observed a significant difference between these three groups of patients, P = 0.026 and P = 0.024, respectively, for nt and aa complexity calculated by Shannon entropy. At baseline, we observed that PCR products from the SVR group displayed more frequently synonymous substitutions (dS) and non-synonymous substitutions (dN) than PCR products from the NR group along the NS5A region, ISDR and PKRbd domains, but not in the V3 domain for dS (Table 3, columns 3 and 4). dN/dS ratios were similar in the two groups in ISDR and PKRbd domains. This ratio was two times higher in SVR patients than in NR patients along the NS5A region and three times higher in the V3 domain (Table 3, column 5).
| Patient andtime of treatment | Sex | Age | Viral loadlog IU/mL | n | NT substitutions/mastersequence | AA substitutions/mastersequence | Genetic distance |
| SVR1-D0 | F | 48 | 2.00 | 15 | 4.5 ± 2.3 | 2.5 ± 1.4 | 0.0065 ± 0.0008 |
| SVR2-D0 | F | 53 | 4.55 | 13 | 5.1 ± 3.6 | 3.3 ± 2.1 | 0.0070 ± 0.0011 |
| SVR3-D0 | M | 43 | 4.71 | 8 | 10.3 ± 3.6 | 4.7 ± 1.9 | 0.0105 ± 0.0018 |
| SVR4-D0 | M | 59 | 6.24 | 16 | 15.4 ± 8.0 | 7.5 ± 4.9 | 0.0172 ± 0.0022 |
| SVR4-M3 | - | - | 1.61 | 13 | 3.6 ± 2.1 | 1.8 ± 1.0 | 0.0051 ± 0.0008 |
| SVR5-D0 | F | 65 | 5.44 | 15 | 10.1 ± 6.6 | 4.9 ± 2.0 | 0.0171 ± 0.0016 |
| SVR5-M3 | - | - | 2.77 | 17 | 6.4 ± 2.4 | 3.3 ± 1.6 | 0.0070 ± 0.0010 |
| SVR6-D0 | M | 47 | 7.26 | 17 | 15.9 ± 10.5 | 8.5 ± 5.1 | 0.0192 ± 0.0022 |
| SVR6-M3 | - | - | 2.18 | 6 | 4.0 ± 1.7 | 2.4 ± 1.3 | 0.0062 ± 0.0012 |
| SVR7-D0 | F | 51 | 6.61 | 15 | 29.3 ± 8.5 | 7.3 ± 2.7 | 0.0294 ± 0.0029 |
| SVR7-M3 | - | - | 5.42 | 15 | 12.6 ± 4.2 | 4.6 ± 2.2 | 0.0156 ± 0.0016 |
| NR1-D0 | M | 35 | 5.13 | 17 | 15.2 ± 4.3 | 3.5 ± 1.9 | 0.0181 ± 0.0020 |
| NR1-M3 | - | - | 5.24 | 9 | 15.3 ± 4.4 | 4.8 ± 1.7 | 0.0143 ± 0.0020 |
| NR1-M6 | - | - | 2.83 | 4 | 4.5 ± 3.1 | 2.0 ± 3.4 | 0.0041 ± 0.0013 |
| NR2-D0 | F | 50 | 5.90 | 17 | 15.6 ± 6.4 | 5.2 ± 2.9 | 0.0181 ± 0.0020 |
| NR2-M3 | - | - | 5.87 | 12 | 9.5 ± 3.6 | 3.9 ± 1.8 | 0.0125 ± 0.0013 |
| NR2-M6 | - | - | 5.77 | 10 | 10.3 ± 3.3 | 4.0 ± 2.2 | 0.0274 ± 0.0020 |
| NR3-D0 | F | 44 | 5.63 | 14 | 10.9 ± 3.7 | 4.4 ± 2.2 | 0.0279 ± 0.0021 |
| NR3-M3 | - | - | 5.79 | 11 | 11.1 ± 2.7 | 4.0 ± 2.8 | 0.0157 ± 0.0017 |
| NR3-M6 | - | - | 5.00 | 12 | 10.8 ± 3.3 | 4.4 ± 2.0 | 0.0154 ± 0.0015 |
| Region andpatient type | Geneticdistance | dS | dN | dN/dS | Num. amino acidmutations |
| NS5A | |||||
| SVR (n = 99) | 0.1027 | 0.3239 | 0.039 | 0.120 | 33.9 |
| NR (n = 48) | 0.0698 | 0.1216 | 0.025 | 0.206 | 29.8 |
| ISDR | |||||
| SVR (n = 99) | 0.0834 | 0.3084 | 0.0295 | 0.096 | 1.3 |
| NR (n = 48) | 0.0765 | 0.2645 | 0.0271 | 0.102 | 1.9 |
| PKRbd | |||||
| SVR (n = 99) | 0.0934 | 0.2934 | 0.0424 | 0.145 | 5.3 |
| NR (n = 48) | 0.0815 | 0.2694 | 0.0332 | 0.123 | 5.7 |
| V3 | |||||
| SVR (n = 99) | 0.1836 | 0.3264 | 0.1282 | 0.393 | 6.0 |
| NR (n = 48) | 0.1372 | 0.4482 | 0.0556 | 0.124 | 4.7 |
Among the aa sequences of the PKRbd and V3 domains, we observed that NR patients exhibited a higher variant complexity than SVR patients before initiation of IFN therapy in the three domains ISDR, PKRbd and V3 (Table 4). In ISDR, after three months of IFN therapy, two out of four slow SVR patients and the three NR patients showed a decrease in variant complexity (SVR5; SVR6; NR1; NR2 and NR10). The two other slow SVR patients displayed stability in variant complexity (SVR4 and SVR7). The three NR patients had a continuous decrease in the complexity under combined IFN-R regimen at M6 (NR1; NR2 and NR3).
| Patient | Time of treatmentdomains | D0ISDR-PKRbd-V3 | M3ISDR-PKRbd-V3 | M6ISDR-PKRbd-V3 |
| SVR1 | n = 15 | 3–4–3 | - | - |
| SVR2 | n = 13 | 3–4–2 | - | - |
| SVR3 | n = 8 | 3–5–3 | - | - |
| SVR4 | n = 16–13 | 3–7–3 | 3–4–3 | - |
| SVR5 | n = 15–17 | 8–8–1 | 4–7–1 | - |
| SVR6 | n = 17–6 | 8–9–3 | 1–2–3 | - |
| SVR7 | n = 15–17 | 6–6–7 | 6–7–7 | - |
| NR1 | n = 17–9–4 | 6–8–7 | 3–4–4 | 1–3–2 |
| NR2 | n = 17–12–10 | 7–8–2 | 5–8–3 | 3–4–6 |
| NR3 | n = 14–11–12 | 6–7–4 | 5–7–3 | 4–5–2 |
Within PKRbd, after three months of IFN therapy, three out of four slow SVR patients and one out of three NR patients displayed a decrease in variant complexity (SVR4; SVR5; SVR6 and NR2) (Table 4). The two other NR patients exhibited stability in variant complexity (NR2 and NR3) and the fourth slow SVR patient showed an increase in variant complexity (SVR7). The three NR patients had a decrease in complexity under combined IFN-R therapy at M6.
After three months of IFN therapy, three out of four slow SVR patients (SVR4; SVR6 and SVR7) displayed a decrease in variant complexity in the V3 domain, as did two out of three NR patients (NR1 and NR3) (Table 4). The two other patients were found to contain an increase in variant complexity (SVR5 and NR2). Under combined IFN-R therapy, two NR patients had a low decrease in variant complexity (NR1 and NR3), whilst the third NR patient showed an increase (NR2) in variant complexity.
Comparison to master sequence: Among the 256 PCR products studied, before and during treatment, we observed a mean of 11.7 ± 7.9 nt substitutions with a median of 10 nt substitutions.
At baseline, we observed 13.6 ± 9.1 nt substitutions (median of 12 nt substitutions) among the 147 clones analyzed, in comparison to the master sequence (Table 2, column 6). The aa variations were 4.8 ± 3.1 substitutions (with a median at 4) among all 256 clones and 5.4 ± 3.5 aa substitutions in clones analyzed before treatment, with a median of 5 aa substitutions. We also observed that all variants from SVR patients had a higher genetic diversity than all variants from NR patients in the four regions studied, including ISDR, PKRbd, V3 domain and complete NS5A (Table 3, column 2). Within patients, we observed that the 3 early SVR had a lower genetic diversity than the four slow SVR and the three NR.
We observed significantly more aa substitutions in SVR clones than in NR clones in two out of six regions (complete NS5A, P = 0.013; the first half of NS5A, P = 0.032). Interestingly, NR clones presented more aa substitutions in the V3 domain than SVR clones, but this was not statistically significant (Table 3).
During treatment, the genetic distance and numbers of substitutions (nt and aa) in SVR PCR products decreased between the baseline and M3 (Table 2, columns 6, 7 and 8). PCR products from NR patients also showed a decrease of the genetic distance, but this was lower than those from SVR patients. However, clones from NR patients had either stability or an increase in the number of aa substitutions observed under treatment. NS5A variants had a high heterogeneity without clonal selection of quasispecies and a low variation in ISDR and V3 domains during therapy.
Comparison to HCV-J sequence: Among the 256 clones analyzed, we observed a mean of 32.1 ± 5.2 aa substitutions. At baseline, a mean of 36.6 ± 5.6 aa substitutions (median at 32 aa substitutions) was observed among the 147 clones analyzed, in comparison to the HCV-J sequence (Tables 2 and 5). Next, we determined for each group the level of aa substitutions in each site, with a threshold at 15 aa substitutions (this threshold was defined as half of the maximum (n = 31) of aa substitutions observed for the 256 clones; in position 2218). In ISDR, aa variations were found at positions 2218, 2224, 2232, 2234 and 2237, in the V3 domain at position 2377 and along NS5A protein at positions 2251 (PKRbd), 2280 and 2408.
| NS5A region (aa range) | 2 groups (D0)(n = 147) | SVR (D0)(n = 99) | NR (D0)(n = 48) | SVR (M3)(n = 51) | NR (M3)(n = 32) | NR (M6)(n = 26) |
| ISDR (2209-2248) | 1.5 ± 1.6 | 1.3 ± 1.7 | 1.9 ± 1.1 | 0.8 ± 0.8 | 2.1 ± 1.0 | 1.8 ± 1.2 |
| PKRbd (2209-2274) | 5.4 ± 2.1 | 5.3 ± 2.4 | 5.7 ± 1.2 | 4.7 ± 1.6 | 6.1 ± 1.0 | 5.7 ± 1.3 |
| V3 (2353-2379) | 5.5 ± 1.4 | 6.0 ± 1.5 | 4.7 ± 0.6 | 6.1 ± 1.3 | 4.8 ± 0.7 | 5.1 ± 0.6 |
| Complete NS5A (1973-2208) | 32.6 ± 5.6 | 33.9 ± 6.0 | 29.8 ± 3.7 | 31.7 ± 5.3 | 30.8 ± 3.9 | 31.9 ± 2.9 |
| N-terminal part of NS5A (1973-2208) | 12.4 ± 3.1 | 12.9 ± 3.3 | 11.6 ± 2.4 | 11.7 ± 2.0 | 11.2 ± 2.1 | 12.8 ± 2.1 |
| C-terminal part of NS5A (2209-2419) | 20.1 ± 3.9 | 21.1 ± 4.1 | 18.2 ± 2.4 | 20.1 ± 4.2 | 19.6 ± 2.9 | 19.4 ± 3.4 |
At baseline, among the two group of patients (SVR and NR), the comparison of the different clone populations showed that all except one (PKRbd) of the six regions studied were able to differentiate the two groups (Table 5).
We observed more aa substitutions in SVR clones than in NR clones in four out of six regions (complete NS5A, P < 0.001; the first half of NS5A, P = 0.010; the second half of NS5A, P < 0.001 and V3, P < 0.001). Interestingly, NR clones presented more aa substitutions in ISDR than SVR clones, P = 0.011, and the PKRbd domain was not able to discriminate the two groups of patients. In contrast to previous findings, NR clones had more aa substitutions than SVR clones in ISDR and PKRbd regions. We did not find any correlation between HCV viral load and the number of mutations observed in ISDR and the V3 domains. A fair correlation was observed between HCV viral load and PKRbd (r = 0.724; P = 0.018). A good correlation was observed between HCV viral load and complete NS5A (r = 0.855; P = 0.002) and with the second half of NS5A (r = 0.782; P = 0.008).
During treatment, a decrease of aa substitutions occurred in the six studied regions for the two groups of patients. Only ISDR and the complete NS5A sequence showed a significant decrease in aa substitutions under treatment in comparison with populations studied at baseline (P = 0.019 and P = 0.005, respectively). In the NR clones, stability or a low decrease in aa substitutions was observed in the six regions, at baseline, M3 and M6.
Among the three populations of clones from NR patients, we did not find any aa substitutions between M3 and M6 treatment points, as recently described in NS5A during ribavirin monotherapy[38]. No other specific aa substitutions were found in the NS5A protein.
Numerous viral and host factors have been described as predictors of response to therapy. HCV genotype 1, particularly genotype 1b, are most resistant to combined IFN-R therapy[5]. Among viral factors, genetic variability has been studied in many regions of the HCV genome, mainly in hypervariable region 1 (HVR1) and PKR-eIF2α phosphorylation homology domain (PePHD) of E2 region and in PKRbd (including ISDR) of NS5A[17,22,39-43]. However, few reports have studied the complete NS5A region[17,22]. Most of these studies examined patients treated by IFN monotherapy, with few studies of patients treated by combined IFN-R therapy.
A recent study examining viral sequence differences between African American and Caucasian patients treated by IFN with or without ribavirin found that the NS5A region did not cluster by race, but treatment response and IFN effectiveness did[44]. In this study, our analysis of the number of substitutions in the V3 domain showed a significant difference between the two groups of patients and a significant correlation between IFN effectiveness and a high number of mutations in the V3 domain[45].
In another study examining the second half of the NS5A region, especially ISDR, PKRbd and V3 domains, in patients treated by combined IFN-R therapy, the authors also found that the V3 domain showed more accumulation of substitutions than other domains in NS5A[45]. SVR patients had lower complexity and diversity than NR patients before treatment. During therapy, the investigators observed a decrease in complexity and diversity in SVR patients, and an increase in complexity with accumulation of non-synonymous mutations in NR patients.
In our study, we analyzed sequence variations of NS5A quasispecies in patients who received adapted therapy according to virological response during treatment. Among SVR patients, we analyzed three early SVR and four slow SVR patients compared to three NR patients. The construction of a phylogenetic tree did not allow us to distinguish the two groups of patients (SVR and NR). Before treatment, the analysis of genetic complexity and diversity did not show a significant difference between the seven SVR and the three NR patients. However, the genetic complexity and diversity were lower in early SVR than in the seven other SVR patients. The analysis of the immunity pressure in different domains of the NS5A gene did not show differences between the SVR and NR groups except for the V3 domain and the complete NS5A region where the SVR group presented a higher immunity pressure than NR patients. Generally, the analysis of mutations compared to the master sequence or HCV-J sequences requires long fragments (a half or a complete NS5A protein) to distinguish SVR and NR. We did not observe any difference in specific aa substitutions between the two groups of patients.
During treatment, we observed a decrease of genetic complexity and diversity in slow SVR and NR patients. A very low complexity was found in the V3 domain in eight out of ten patients (6/7 of SVR and 2/3 of NR patients). In the PKRbd and V3 domains, we often observed complexity decrease or stability. Only one out of seven SVR and one out of three NR patients displayed complexity increase either under IFN therapy or under combined IFN-R therapy. Thus, we observed a different evolution of quasispecies under IFN alone or under combined therapy. This is in contrast to a recent study by Puig-Basagoti et al[45] where they observed a decrease in quasispecies complexity in SVR patients and a stability or increase in quasispecies complexity in NR patients. In this study, the analysis of the quasispecies was done at wk 1, 2 and 4, whereas in our study we analyzed quasispecies after a first three-month period of treatment under IFN alone and after a second three-month period under combined IFN-R therapy. We observed the same rate of appearance of aa substitutions in the ISDR domains for the two categories of patients. However, the instauration of a real modification of aa in a quasispecies was more often observed in SVR patients than in NR patients in the complete NS5A protein and in the PKRbd and V3 domains. Interestingly, in three slow SVR (SVR4-SVR5-SVR6) patients the main variant from the ISDR domain remained unchanged after three months of treatment. The same observation was made in two out of three NR patients who also exhibited complexity stability in the different NS5A regions.
Asahina et al[38] studied the possible mutagenic effect of ribavirin during a period of 28 days of ribavirin monotherapy before combined IFN-R therapy. During the first phase of ribavirin monotherapy, they observed an accumulation of non-synonymous substitutions located in NS5A. These substitutions were observed in ten patients and eight of them had SVR after treatment. In our study, we did not observe these substitutions in PKRbd nor in any part of NS5A. However, our treatment regimen was different and we only studied the effect of ribavirin during three months of combined therapy in NR patients. It is also possible that substitutions did arise during combined therapy, but were subsequently eliminated by the treatment.
In conclusion, our study reinforces the potential role of NS5A polymorphism, particularly in the V3 domain, in regulating HCV resistance to IFN treatment. Nevertheless, the underlying molecular mechanisms remain unclear. Mutations in the V3 domain may affect the interaction between NS5A and one or several IFN-induced antiviral effectors. Some studies found high mutation frequency in ISDR being associated with low HCV RNA titer, suggesting that NS5A may play a role in HCV replication. In our study, at baseline, we did not find any relationship between HCV viral load and the number of mutations observed in the ISDR and V3 domains. The number of mutations observed in PKRbd has shown a fair correlation with HCV viral load, but we found a better correlation when we analyzed the second half or the complete NS5A region. These results support the notion that mutations in the NS5A region may influence on HCV replication, and suggest that the entire NS5A region should be analyzed for mutations that may correlate with IFN response. Further clinical studies with new therapeutic approaches such as pegylated IFN, as well as the use of the recently developed HCV infection cell culture systems should further improve our understanding of the role of NS5A in regulation of the IFN response.
The authors thank Schering-Plough for supporting the therapeutic protocol and all hepatologists and virologists of the Group of Fontevraud who participated to this therapeutic protocol. The authors thank Kevin L Erwin for his careful proofreading of the manuscript.
In this article, we analyzed NS5A HCV quasispecies evolution during antiviral treatment. Since the NS5A protein is implicated in HCV resistance, this study should enhance the knowledge of evolution of the resistance during treatment.
Many previous articles focused on a small region(s) of NS5A without studying the complete gene. We believe that an analysis of the entire NS5A region is necessary because some sections of NS5A may be important for homologous and/or heterologous RNA-protein or protein-protein interactions and thus may play a role in conferring HCV resistance to IFN therapy.
To the best of our knowledge, only two reports studied quasisipecies along the complete NS5A region and did not observe quasispecies evolution during therapy. In contrast, our results suggest that V3 domain plays a role in HCV resistance to IFN therapy.
Our study may help identify mutations within the HCV genome that can be used to predict patients’ response to IFN treatment or future new antiviral modalities, as well as provide molecular hints for the mechanisms underlying HCV resistance to the therapy.
Study is important with findings suppporting previous results as well as novel findings. The biggest problem is the very small size of the patients analyzed. The paper is well written and consise.
S- Editor Liu Y L- Editor Zhu LH E- Editor Lu W
| 1. | Alberti A, Chemello L, Benvegnù L. Natural history of hepatitis C. J Hepatol. 1999;31 Suppl 1:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 239] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 2. | Zeuzem S, Hultcrantz R, Bourliere M, Goeser T, Marcellin P, Sanchez-Tapias J, Sarrazin C, Harvey J, Brass C, Albrecht J. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J Hepatol. 2004;40:993-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 254] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 3. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4736] [Cited by in RCA: 4558] [Article Influence: 189.9] [Reference Citation Analysis (0)] |
| 4. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4847] [Cited by in RCA: 4747] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 5. | Poynard T, McHutchison J, Goodman Z, Ling MH, Albrecht J. Is an "a la carte" combination interferon alfa-2b plus ribavirin regimen possible for the first line treatment in patients with chronic hepatitis C? The ALGOVIRC Project Group. Hepatology. 2000;31:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 250] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 6. | Hofmann WP, Zeuzem S, Sarrazin C. Hepatitis C virus-related resistance mechanisms to interferon alpha-based antiviral therapy. J Clin Virol. 2005;32:86-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Gale M, Katze MG. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 311] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 8. | Tan SL, Gale MJ, Katze MG. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol Cell Biol. 1998;18:2431-2443. [PubMed] |
| 9. | Gale MJ, Korth MJ, Katze MG. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol. 1998;10:157-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 113] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 452] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 11. | Arase Y, Ikeda K, Chayama K, Murashima N, Tsubota A, Suzuki Y, Saitoh S, Kobayashi M, Kobayashi M, Kobayashi M. Efficacy and changes of the nonstructural 5A GENE by prolonged interferon therapy for patients with hepatitis C virus genotype 1b and a high level of serum HCV-RNA. Intern Med. 1999;38:461-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 12. | Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 152] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 13. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 733] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 14. | Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 124] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Nakano I, Fukuda Y, Katano Y, Nakano S, Kumada T, Hayakawa T. Why is the interferon sensitivity-determining region (ISDR) system useful in Japan? J Hepatol. 1999;30:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Berg T, Mas Marques A, Höhne M, Wiedenmann B, Hopf U, Schreier E. Mutations in the E2-PePHD and NS5A region of hepatitis C virus type 1 and the dynamics of hepatitis C viremia decline during interferon alfa treatment. Hepatology. 2000;32:1386-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373-1381. [PubMed] |
| 18. | Sarrazin C, Berg T, Lee JH, Rüster B, Kronenberger B, Roth WK, Zeuzem S. Mutations in the protein kinase-binding domain of the NS5A protein in patients infected with hepatitis C virus type 1a are associated with treatment response. J Infect Dis. 2000;181:432-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Squadrito G, Orlando ME, Cacciola I, Rumi MG, Artini M, Picciotto A, Loiacono O, Siciliano R, Levrero M, Raimondo G. Long-term response to interferon alpha is unrelated to "interferon sensitivity determining region" variability in patients with chronic hepatitis C virus-1b infection. J Hepatol. 1999;30:1023-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Zeuzem S, Lee JH, Roth WK. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Chung RT, Monto A, Dienstag JL, Kaplan LM. Mutations in the NS5A region do not predict interferon-responsiveness in american patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 22. | Nousbaum J, Polyak SJ, Ray SC, Sullivan DG, Larson AM, Carithers RL, Gretch DR. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J Virol. 2000;74:9028-9038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 23. | Murphy MD, Rosen HR, Marousek GI, Chou S. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig Dis Sci. 2002;47:1195-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Pascu M, Martus P, Höhne M, Wiedenmann B, Hopf U, Schreier E, Berg T. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut. 2004;53:1345-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Podevin P, Sabile A, Gajardo R, Delhem N, Abadie A, Lozach PY, Beretta L, Bréchot C. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology. 2001;33:1503-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Inchauspe G, Zebedee S, Lee DH, Sugitani M, Nasoff M, Prince AM. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci USA. 1991;88:10292-10296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 182] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 27. | Liu S, Ansari IH, Das SC, Pattnaik AK. Insertion and deletion analyses identify regions of non-structural protein 5A of Hepatitis C virus that are dispensable for viral genome replication. J Gen Virol. 2006;87:323-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Appel N, Pietschmann T, Bartenschlager R. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol. 2005;79:3187-3194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 29. | Lunel F, Veillon P, Fouchard-Hubert I, Loustaud-Ratti V, Abergel A, Silvain C, Rifflet H, Blanchi A, Causse X, Bacq Y. Antiviral effect of ribavirin in early non-responders to interferon monotherapy assessed by kinetics of hepatitis C virus RNA and hepatitis C virus core antigen. J Hepatol. 2003;39:826-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495-503. [PubMed] |
| 31. | Payan C, Véal N, Crescenzo-Chaigne B, Bélec L, Pillot J. New quantitative assay of hepatitis B and C viruses by competitive PCR using alternative internal sequences. J Virol Methods. 1997;65:299-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 32. | Malet I, Belnard M, Agut H, Cahour A. From RNA to quasispecies: a DNA polymerase with proofreading activity is highly recommended for accurate assessment of viral diversity. J Virol Methods. 2003;109:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876-4882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29832] [Cited by in RCA: 27618] [Article Influence: 986.4] [Reference Citation Analysis (0)] |
| 34. | Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 447] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 35. | Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20000] [Cited by in RCA: 18574] [Article Influence: 412.8] [Reference Citation Analysis (0)] |
| 36. | Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418-426. [PubMed] |
| 37. | Gaudy C, Moreau A, Veillon P, Temoin S, Lunel F, Goudeau A. Significance of pretreatment analysis of hepatitis C virus genotype 1b hypervariable region 1 sequences to predict antiviral outcome. J Clin Microbiol. 2003;41:3615-3622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 38. | Asahina Y, Izumi N, Enomoto N, Uchihara M, Kurosaki M, Onuki Y, Nishimura Y, Ueda K, Tsuchiya K, Nakanishi H. Mutagenic effects of ribavirin and response to interferon/ribavirin combination therapy in chronic hepatitis C. J Hepatol. 2005;43:623-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 39. | Gaudy C, Lambelé M, Moreau A, Veillon P, Lunel F, Goudeau A. Mutations within the hepatitis C virus genotype 1b E2-PePHD domain do not correlate with treatment outcome. J Clin Microbiol. 2005;43:750-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Pawlotsky JM, Pellerin M, Bouvier M, Roudot-Thoraval F, Germanidis G, Bastie A, Darthuy F, Rémiré J, Soussy CJ, Dhumeaux D. Genetic complexity of the hypervariable region 1 (HVR1) of hepatitis C virus (HCV): influence on the characteristics of the infection and responses to interferon alfa therapy in patients with chronic hepatitis C. J Med Virol. 1998;54:256-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 41. | Polyak SJ, Nousbaum JB, Larson AM, Cotler S, Carithers RL, Gretch DR. The protein kinase-interacting domain in the hepatitis C virus envelope glycoprotein-2 gene is highly conserved in genotype 1-infected patients treated with interferon. J Infect Dis. 2000;182:397-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 42. | Ueda E, Enomoto N, Sakamoto N, Hamano K, Sato C, Izumi N, Watanabe M. Changes of HCV quasispecies during combination therapy with interferon and ribavirin. Hepatol Res. 2004;29:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 43. | Gerotto M, Dal Pero F, Pontisso P, Noventa F, Gatta A, Alberti A. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology. 2000;119:1649-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Layden-Almer JE, Kuiken C, Ribeiro RM, Kunstman KJ, Perelson AS, Layden TJ, Wolinsky SM. Hepatitis C virus genotype 1a NS5A pretreatment sequence variation and viral kinetics in African American and white patients. J Infect Dis. 2005;192:1078-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Puig-Basagoiti F, Forns X, Furcić I, Ampurdanés S, Giménez-Barcons M, Franco S, Sánchez-Tapias JM, Saiz JC. Dynamics of hepatitis C virus NS5A quasispecies during interferon and ribavirin therapy in responder and non-responder patients with genotype 1b chronic hepatitis C. J Gen Virol. 2005;86:1067-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |