Published online Feb 21, 2007. doi: 10.3748/wjg.v13.i7.1018
Revised: December 3, 2006
Accepted: January 18, 2007
Published online: February 21, 2007
AIM: To detect the expression of fragile histidine triad (FHIT) in normal colorectal tissue, colorectal adenoma and colorectal cancer (CRC) tissue, and to analyze its relationship with the clinicopathological features of CRC, and apoptosis-associated proteins (Bcl-2, Bax, survivin) and apoptosis in colorectal cancer.
METHODS: FHIT mRNA analysis was performed by nested reverse transcription-polymerase chain reaction (RT-PCR) assay. Tissue microarray (TMA) was established to detect the expression of FHIT, Bcl-2, Bax and survivin genes in 80 CRC tissue specimens, 16 colorectal adenoma tissue specimens and 16 hemorrhoid (PPH) tissue specimens during the same period of time as the control. Citrate-microwave-SP was used as immunohistochemical method. The relationship between clinicopathological factors, such as differentiation grades and 5-year survival rate was observed. TUNEL assay was used to detect the apoptosis index in 80 CRC tissue specimens.
RESULTS: Ten out of 26 (38.5%) CRC tissue specimens expressed aberrant FHIT transcripts, none of the aberrant FHIT transcripts was observed in the matched normal tissue and colorectal adenoma tissue by nested RT-PCR assay. The positive rate of FHIT gene expression in normal colorectal tissue, colorectal adenoma and carcinoma tissue was 93.75%, 68.75% and 46.25%, respectively. Clinicopathological analysis of patients showed that the decreased FHIT gene expression was not associated with age, sex, serum CEA levels, tumor site and size, histological classification. However, the expression of FHIT was correlated with differentiation grades, pathological stages, lymph node metastases and 5-year survival rate after operation. The positive rate of apoptosis-associated proteins (Bax, Bcl-2 and survivin) in CRC tissue was 72.50%, 51.25% and 77.50%, respectively. The expression of these apoptosis-associated proteins in CRC tissue was correlated with the expression of FHIT. The mean apoptosis index in FHIT negative tumors was significantly lower than that in FHIT positive tumors (5.41 ± 0.23 vs 0.56 ± 0.10, P < 0.01).
CONCLUSION: The FHIT gene plays an important role in the regulation of apoptosis and decreased FHIT expression plays a key role in the initiation and progression of colorectal carcinoma.
- Citation: Cao J, Chen XP, Li WL, Xia J, Du H, Tang WB, Wang H, Chen XW, Xiao HQ, Li YY. Decreased fragile histidine triad expression in colorectal cancer and its association with apoptosis inhibition. World J Gastroenterol 2007; 13(7): 1018-1026
- URL: https://www.wjgnet.com/1007-9327/full/v13/i7/1018.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i7.1018
Colorectal cancer is the second leading cause of death in the United States, where the cumulative lifetime risk of developing colorectal cancer in both men and women is 6%[1]. In China, colorectal cancer is the fourth leading cause of death[2].
The fragile histidine triad (FHIT) gene, is a tumor suppressor gene located at the fragile site FRA3B on chromosome 3p14.2 that can be identified by positional cloning, a region of the genome showing loss of heterozygosity (LOH) in a variety of cancers[3,4]. FHIT is involved in carcinogenesis of many human tissues, including digestive tract tissue. The FHIT gene contains an open reading frame (ORF) of 444 base pairs (bps) encoding a protein of 147 amino acids, which appears to be ubiquitously expressed in human tissues. Large deletions within FHIT transcripts occur frequently in multiple malignancies[5,6]. Most of the deletions described to date in the FHIT gene involve loss of one or more coding exons and lead to a truncated protein[6]. Loss of FHIT protein expression and abnormal FHIT transcripts, including deletions and insertions of exons, have been found in lung and breast cancers, as well as in head and neck cancers[7-11]. Ohta et al[3] reported that aberrant transcripts of the FHIT gene have been observed in 38% of CRC cases. It was also reported that the FHIT gene plays an important role in both oncogenesis and progression of CRC[12-14]. However, Thiagalingam et al[15] showed that there is no evidence that the FHIT gene is involved in CRC carcinogenesis. The clinicopathological significance of FHIT alterations in CRC still remains unclear.
The role of FHIT expression in the development of colorectal cancer (CRC) is poorly understood. Recently, tissue microarray (TMA) technique has been developed, which can significantly increase the throughput of immunohistochemistry (IHC) tumor analysis. TMA is composed of a large number of small tissue cores punched out from different tumors. These tissue core specimens obtained from tissue blocks, are then arranged into a single recipient paraffin block[16]. This approach allows analysis of a large number of different tumor samples in one IHC experiment. Arrays as large as 1000 samples have been reported[17]. TMA can be used to test the prognostic significance of antibodies against proteins encoded by differentially expressed genes using a large number of archival patient samples. Decreased p53 expression is correlated with lymph node metastasis in CRC[18]. In our present study, an expanded repertoire of TMA technique was used to demonstrate the novel relationship between FHIT expression and clinicopathological factors, such as differentiation, pathological stages, lymph node metastasis and 5-year survival rate.
The presence of FHIT gene mutations correlates significantly with decreased FHIT expression in human CRC. Several well integrated biochemical subroutines promote CRC cell growth and inhibit apoptosis, tipping the balance of colon epithelial cells towards an unrestricted proliferation and outliving the sequence of mutations required for carcinogenesis. Apoptosis is essential for successful embryonic development and maintenance of normal tissue homeostasis[19]. The present study was to test the hypothesis that decreased FHIT expression inhibits apoptosis and decreases apoptosis-associated protein expression, to characterize FHIT mRNA and protein expression in normal colorectal tissue, and adenoma and adenocarcinoma tissue, and to investigate the mechanism of FHIT gene inactivation and its association with apoptosis inhibition in the development and progression of human CRC.
Human colorectal tissue specimens were obtained from patients at the Department of Gastrointestinal Surgery, Affiliated Guangzhou First People’s Hospital, Guangzhou Medical College, China, from January 1997 to June 2000. All tissue specimens were snap- frozen and stored at -80°C. There were 48 male and 32 female patients with an average age of 62 years (range 32-81 years). Of the 80 specimens of human colorectal carcinoma we examined, 20, 22, 23 and 15 were classified as Dukes’ stage A, B, C and D, respectively. Histologically, well-differentiated CRC was found in 16, moderately-differentiated CRC in 44, and poorly-differentiated in 22 specimens, respectively (Table 1). As a control, 16 normal colorectal tissue specimens and 16 colorectal adenoma specimens were also obtained. These tissue specimens were snap-frozen in liquid nitrogen, embedded in paraffin and stored at -80°C. Sections of each paraffin block were stained with hematoxylin and eosin.
| n | FHIT expression | χ2 | P | ||
| Positive | Negative | ||||
| Age | |||||
| < 60 | 34 | 16 | 18 | 0.16 | 0.901 |
| ≥ 60 | 46 | 21 | 25 | ||
| Sex | |||||
| M | 42 | 19 | 23 | 0.04 | 0.849 |
| F | 38 | 18 | 20 | ||
| Differentiated | |||||
| Well | 16 | 12 | 4 | 4.84 | 0.028 |
| Moderately | 44 | 20 | 24 | ||
| Poorly | 20 | 5 | 15 | ||
| Dukes | |||||
| A, B | 45 | 27 | 18 | 7.82 | 0.005 |
| C, D | 35 | 10 | 25 | ||
| Lymph node metastasis | |||||
| Positive | 16 | 11 | 5 | 4.07 | 0.006 |
| Negative | 64 | 26 | 38 | ||
| Survival | |||||
| ≥ 5 yr | 35 | 23 | 12 | 5.77 | 0.016 |
| < 5 yr | 18 | 5 | 13 | ||
Total RNA was isolated from 26 colorectal cancer and 26 normal adjacent colonic tissue specimens, and 14 adenoma and 14 normal adjacent colonic tissue specimens with TRIzolTM reagent (Life Technologies). Nested RT-PCR was carried out as described previously[20]. A 24 μL mixture of total RNA (10 μg), 0.4 mmol/L oligo (dT) (New England Biolabs), and QH2O was prepared. After incubation for 10 min at 70°C, the mixture was quickly cooled on ice, and 16 μL of RT-PCR mixture containing 8 μL of 5 × first strand buffer (Invitrogen) (250 mmol/L Tris–HCl pH 8.3, 375 mmol/L KCl, 15 mmol/L MgCl2), 4 μL of 0.1 M DTT (Invitrogen), 2 μL 10 mmol/L deoxynucleotide mixture (dATP, dTTP, dCTP and dGTP, 10 mmol/L each) (Roche), 2 μL of RNase inhibitor (40 U/μL) (Roche) was added. The reaction proceeded in GeneAmp 2400 (Applied Biosystems) at 45°C for 2 min. After 400 U of SuperScript™ RnaseH-reverse transcriptase (200 U/μL) (Invitrogen) was added and incubated for 45 min at 60°C, the reaction was inactivated at 70°C for 15 min and at 94°C for 3 min. cDNA was used as a template in the following RT-PCR.
Mutation screening in aberrant RT-PCR products was performed using an ABI 377 automated sequencer with a pEGM-T vector as described previously[21]. Aberrant migrating bands were directly sequenced after isolation of bands from low melting agarose and purification on columns.
Construction of tissue microarray (TMA): Archival paraffin-embedded, formalin-fixed tissues from 80 Chinese CRC, 16 colonic adenoma and 16 noncancerous normal mucosa patients were collected. Two independent experienced pathologists selected representative areas from each donor tumor block, and then one 1-mm core was taken from a representative area of the tumor and inserted into a recipient paraffin block to create the TMA[6]. One TMA slide contained 112 samples. We employed the TMA technique to explore the expression of FHIT, Bax, Bcl-2 and survivin in colorectal tumor specimens and its correlation with apoptosis status by TMA-IHC and TMA-TUNEL. We investigated consecutively-cut serial sections and examined the same tumor region in three dimensions.
Immunohistochemical analysis of FHIT, Bax, Bcl-2 and survivin expression in human CRC by TMA-IHC: Immunohistochemical studies were performed using a citrate-microwave-streptavidin peroxydase technique. Immunohistochemical detection was done using anti-FHIT, anti-Bax, anti-Bcl-2 and anti-survivin antibodies, respectively. Endogenous peroxidase activity was quenched by methanol containing 3% hydrogen peroxide (Sigma, Taufkirchen, Germany). Nonspecific binding was blocked by applying normal rabbit serum in a humidity chamber at a dilution of 1:10 for 30 min. Primary antibodies were applied overnight at 4°C. The secondary antibody (goat to mouse immunoglobulins, DAKO, Denmark) was applied for 1 h at room temperature. Peroxidase–antiperoxidase (PAP rabbit, DAKO) conjugate diluted at 1:100 in phosphate-buffered saline (PBS) was applied for 45 min at room temperature. The sections were stained with diaminobenzidine tetrahydrochloride (DAB, Sigma) and then counterstained with hematoxylin. Negative control staining was performed by omitting the primary antibody. As a positive control for FHIT protein in immunohistochemical studies, we used paraffin sections of lung cancer positive for FHIT protein.
Quantitative measurements: Quantitative analysis of immune reactions related to the total tissue area was performed. We measured the percentage of positive immune reactivity with these antibodies in the tissue epithelium (normal and tumor). Histological images were captured with an OLYMPUS BX50 system microscope with an objective at magnification X 40, through a video camera, and digitized by appropriate software. In each section, 500 cells per core were calculated. The relative level of specific immunostaining and its localization were also judged. The relative intensity of cell immunostaining was evaluated semi-quantitatively. The data of tissue array were confirmed by a pathologist at the Department of Pathology of Affiliated Guangzhou First People’s Hospital.
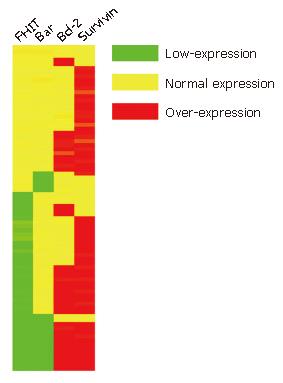
Hierarchical clustering and tree-view analysis: Hierarchical clustering was performed by the Cluster program (available at http://rana.lbl.gov/) as described previously in two dimensions: tumors were grouped together based on the relation of their immunostaining profiles, and antibodies were grouped based on tumors they stained. The clustered data were visualized with the Tree-view software tool programs originally developed for analyzing cDNA microarray data (Available at http://rana.lbl.gov/), which graphically displayed the results of the analysis as dendrograms and arrays, wherein the rows and columns corresponded to the raw staining data, presented in the order determined.
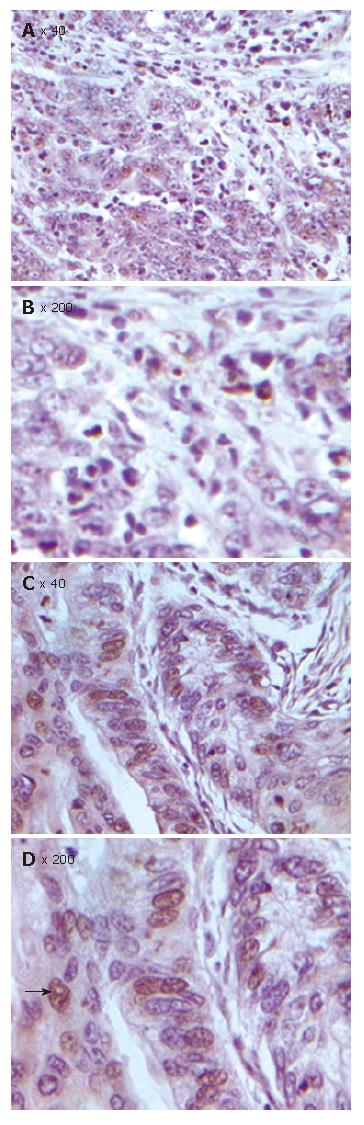
Cell apoptosis was detected in 80 CRC specimens by TMA-TUNEL assay (Apotag Peroxidase Kit, Oncor, Gaithersburg, MD) with the Apop-TagTM peroxidase kit (Zhongshan Biotech, Beijing). For the evaluation of TUNEL index, the number of TUNEL-positive colonic epithelial cells was recorded using the × 40 objective lens. The case was considered positive for TUNEL if any colonic epithelial cells showed TUNEL staining. The TUNEL index was determined as the number of TUNEL-positive colonic epithelial cells expressed as a percentage of the total number of counted colonic epithelial cells. Necrotic areas were excluded.
Results were presented as mean ± SD. All statistical analyses were carried out using SPSS 11.0 for Windows statistical software (SPSS Inc, USA). The relationship between the clinicopathological variables and FHIT loci and protein alterations was examined by Fisher’s exact test. The relationship between the expression of FHIT and prognosis of CRC was analyzed by Cox-Mantel test. P < 0.05 was considered statistically significant. All experiments were performed three times.
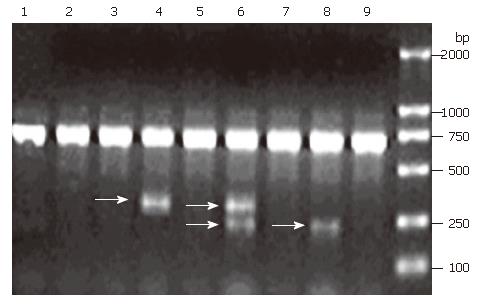
Expression of FHIT mRNA was detected by nested RT-PCR. According to the results, 34.6% samples with abnormal FHIT expression displayed two transcript categories: normal FHIT transcript (PCR products = 707 bp) and aberrant FHIT transcript (PCR products = 336 bp and 239 bp), respectively (Figure 1). However, the samples with normal FHIT expression only showed a normal FHIT transcript size.
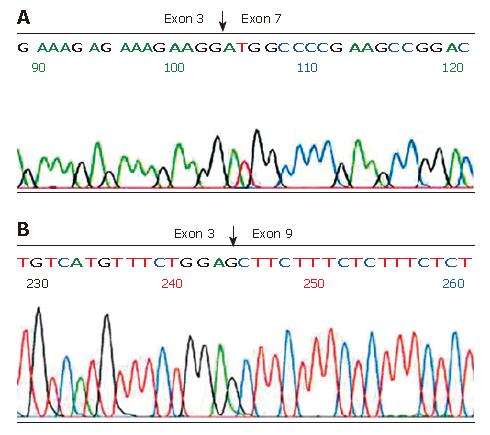
Aberrant RT-PCR products were sequenced after isolation of bands from low melting agarose and purification on columns. In the 336 bp fragment, Exon 3 was spliced to exon 7 (E3/E7), and thus the transcript lacked exons 4-6 (Figure 2A). In the 239 bp fragment, Exon 3 was spliced to exon 9 containing an E3/E9 aberrant transcript (Figure 2B).
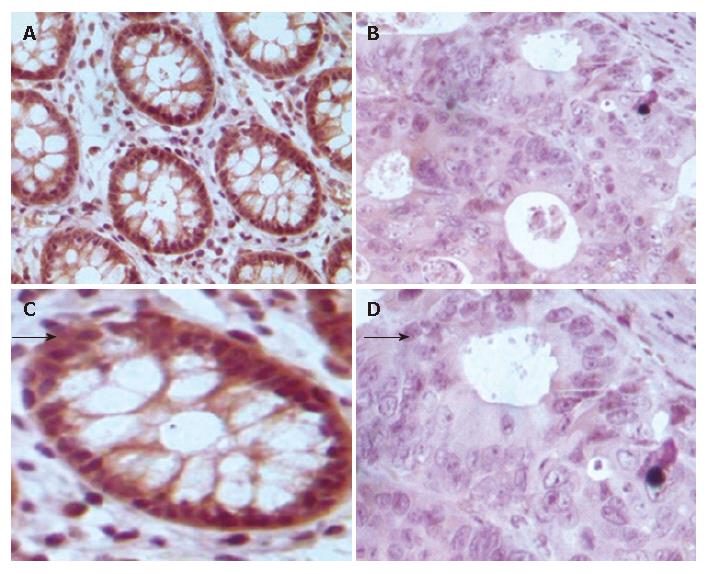
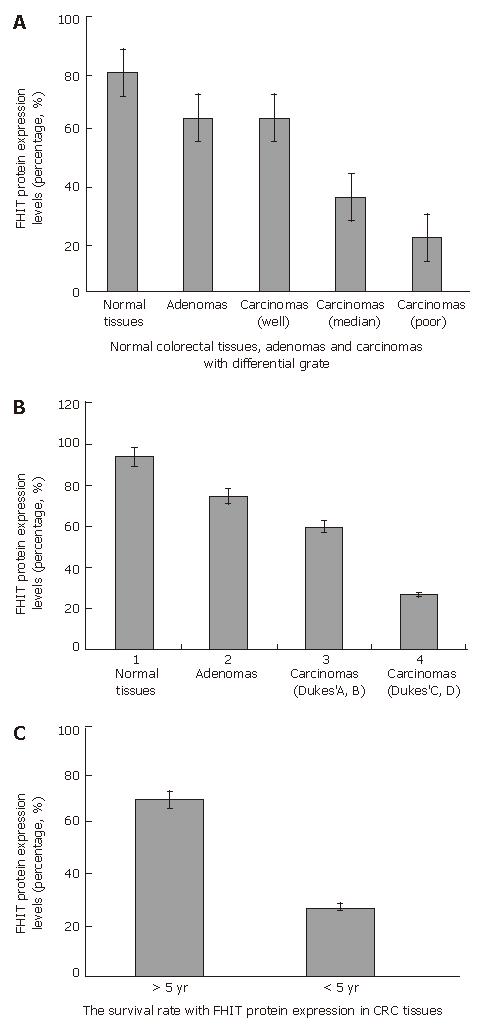
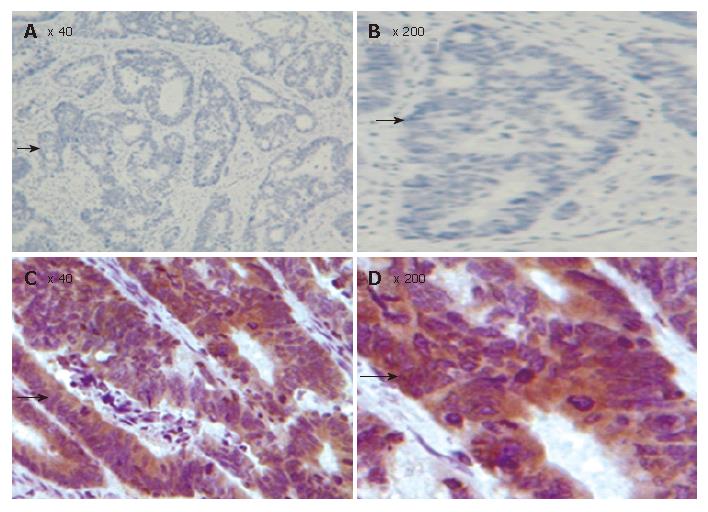
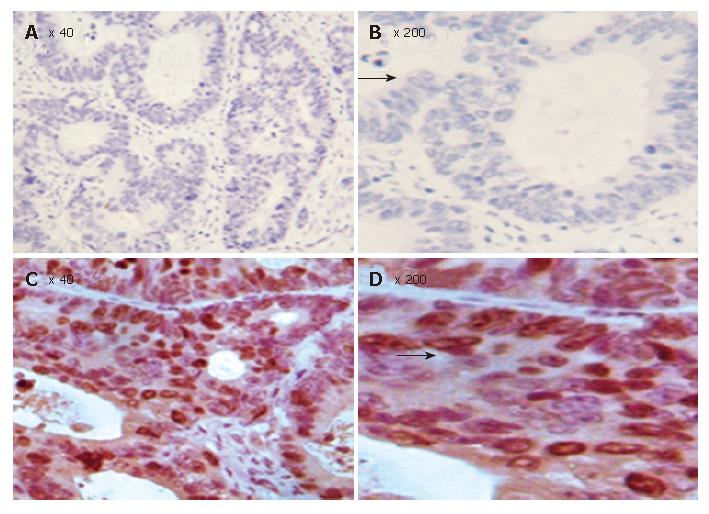
The correlation between FHIT protein expression and clinicopathological data in the 80 carcinoma specimens is shown in Table 1, and a photograph of a representative specimen is provided in Figure 3. FHIT negative CRC was found in 43 and FHIT FHIT positive CRC was found in 37 specimens. Fifty-three percent (32 of 60) of specimens with well-differentiated CRC had positive FHIT expression, whereas 25% (5 of 20) of specimens with poorly-differentiated CRC had positive FHIT expression (P < 0.05), while 93.75% (15 of 16) and 75% (12 of 16) had positive FHIT expression in the normal colorectal tissue specimens and colorectal adenoma specimens, respectively (Figure 4A and Table 1). Sixty percent (27 of 45) of specimens with Dukes’ A and B had positive FHITexpression, whereas only 28% (10 of 35) of specimens with Dukes’ C and D had positive FHIT expression (P < 0.005) (Figure 4B and Table 1). Moreover, prognosis of the FHIT-negative cases was much poorer than the FHIT-positive cases (Figure 4C and Table 1). There was no significant correlation between other clinicopathological factors and FHIT expression.
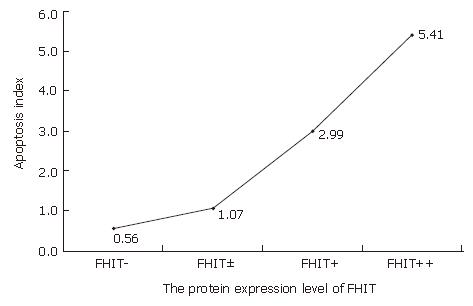
The role of FHIT protein in apoptosis as a proapoptic factor was detected by TUNEL assay. Aberrant expression of the FHIT gene was related to colonic epithelial cell apoptosis (Figures 5 and 6, Table 2). The apoptosis index was 5.41 ± 0.23 in colorectal cancer with normal FHIT expression and 0.56 ± 0.10 in tumors with absent FHIT expression. The rate of apoptosis was significantly lower in tumors with aberrant FHIT expression than in tumors with normal FHIT expression (P < 0.05).
| FHIT | n | AI |
| - | 11 | 0.56 ± 0.10 |
| +/- | 15 | 1.07 ± 0.16 |
| + | 17 | 2.99 ± 0.32 |
| ++ | 37 | 5.41 ± 0.23 |
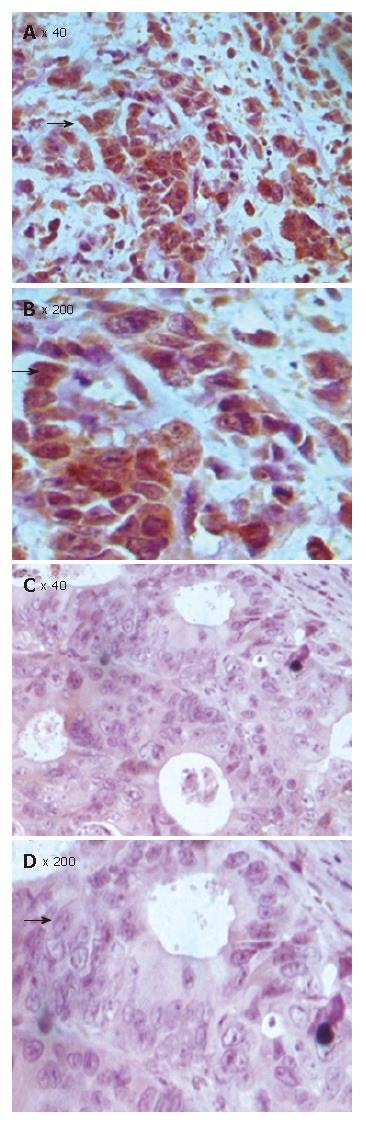
Since growth inhibitory effect of FHIT- expressing cells is related to apoptosis, we determined whether FHIT protein is related with other members in the apoptotic pathway. Of the CRC tissue specimens, 54% (43 of 80) had positive FHIT expression, whereas the remaining 46% (37 of 80) had no detectable FHIT expression by TMA-IHC assay (Figure 7, Figure 8, Figure 9 and Figure 10). Seventy-seven percent of FHIT protein-negative CRC specimens showed decreased Bax expression, whereas 75% and 60% FHIT protein-negative specimens displayed increased Bcl-2 and survivin expression (Figure 7, Figure 8, Figure 9 and Figure 10). The difference was statistically significant (Table 3).
| n | FHIT expression | χ2 | P | |||
| Positive | Negative | |||||
| Bax | + | 62 | 25 | 37 | 3.89 | 0.048 |
| - | 18 | 12 | 6 | |||
| Bcl-2 | + | 58 | 32 | 26 | 6.75 | 0.009 |
| - | 22 | 5 | 17 | |||
| Survivin | + | 41 | 11 | 30 | 12.76 | 0 |
| - | 39 | 26 | 13 | |||
In the present study, frequent allelic loss was observed on chromosome 3p in lung cancer and preneoplastic bronchial lesions, indicating that inactivation of putative tumor suppressor genes on chromosome 3p may be involved in early steps of lung carcinogenesis. Ohta et al[3] have identified the FHIT gene on chromosome 3p14.2, spanning the FRA3B common fragile site and the t (3:8) break point associated with hereditary renal cell carcinomas. The FHIT gene is more than 1 Mb in size, encoding a 1.1-kb cDNA with 10 small exons and a cytoplasmic Mr 16 800 protein with diadenosine triphosphate (Ap3A) hydrolase activity [4]. In this study, truncated FHIT transcripts were observed frequently alongside full-length transcripts and sequence analysis of the truncated gene transcripts revealed mainly exon skipping and alternate RNA processing events. To elucidate the possible molecular mechanisms responsible for this loss of protein expression, we used RT-PCR for FHIT transcript analysis and cDNA sequencing for mutation detection. We found that 34.6% of the samples with decreased FHIT expression had an additional aberrant FHIT transcript product. However, aberrant FHIT was not identified in the samples with normal FHIT expression. These data suggest that loss and rearrangement of the FHIT gene derived from FRA3B breaks or gaps in the large FHIT intronic region may give rise to aberrant cDNA splicing and result in loss of FHIT protein, suggesting that the majority of aberrant FHIT transcripts lack exons 4-6 and 4-8. Our results indicate that higher incidence of aberrant processing of FHIT mRNA and mutation result in FHIT down-regulation in human CRC.
Loss of FHIT protein expression has been found in lung and breast cancers, as well as in head and neck cancers[7-10]. Ohta and colleagues[2] have reported significant loss or reduction of FHIT expression in 39% CRC patients. However, Thiagalingam et al[15] showed that the FHIT gene is not involved in CRC carcinogenesis. In this study, we demonstrated that loss of FHIT expression was associated with poorly- and well-differentiated CRC and frequent lymph node metastasis by TMA-IHC assay, indicating that FHIT plays a role in the progression of CRC because loss of FHIT protein is intimately associated with the development of CRC. Lack of FHIT gene expression and consequent protein expression can be due to deletions, mutations, or epigenetic modifications.
These results strongly indicate that aberrant expression of the FHIT gene is related to the development and progression of CRC. However, the relationship between FHIT expression and apoptosis has not been studied in human CRC, and the decreased apoptosis in FHIT deficient CRC has not been explained. We therefore designed this study to investigate the differential patterns of apoptosis-related protein expression associated with difference in FHIT expression, in order to identify specific changes that might reflect the pathway by which FHIT affects apoptosis. We performed TUNEL analysis in this study to assess apoptosis status and observed decreased apoptosis in FHIT negative CRC tissue samples compared to FHIT-positive CRC tissue samples. The apoptosis index was 5.41 ± 0.23 in tumors with normal FHIT expression and 0.56 ± 0.10 in tumors with aberrant expression. The rate of apoptosis in tumors with aberrant FHIT expression was significantly lower than that in tumors with normal FHIT expression (P < 0.01). Furthermore, we conducted tissue microarray analysis to identify differentially expressed proteins in relevant apoptosis pathways. For this initial pathway, we chose three key proteins belonging to the Bcl/Bax pathway (Bcl2 and Bax) and survivin-caspase pathway (survivin). In this study, the FHIT-negative CRC tissue samples were associated with down-regulation of Bax and up-regulation of Bcl-2. It is known that the Bax gene is an apoptosis-promoting member of the BCL-2 gene family, and apoptosis is known to be accelerated when the Bax function is predominant[22]. On the other hand, over-expression of Bcl-2 seems to be associated with the blocking of apoptosis. The ability of Bcl-2 to inhibit apoptosis is dependent on the expression of Bcl-2 and formation of hetero- and homo-dimers between members of the Bcl-2 family[23]. We observed a significant correlation between FHIT protein expression and members of the Bcl/Bax pathway in human CRC (P < 0.05). Survivin is minimally expressed in normal adult tissues but over-expressed in a wide range of human cancers, including cancer of the breast, lung, esophagus and urinary bladder. Survivin is one member of inhibitors of apoptosis and appears to block apoptosis by interfering with the caspase activation pathway[24]. In the present study, we demonstrated that survivin, an inhibitor of apoptosis protein family, was significantly increased in FHIT-negative CRC, indicating that FHIT plays a critical role in FHIT-induced apoptosis, occurring through inactivation of the survivin-caspase signal pathway. Accordingly, future investigations must address protein-protein interactions that lead to these changes in expression and apoptosis, thus allowing us to place FHIT at its correct position within the pathways, ultimately helping to elucidate the precise mechanism by which its expression modulates apoptosis in human CRC.
In conclusion, decreased FHIT expression is caused by mutations of the FHIT gene in human CRC. Down-regulation of FHIT is associated with down-regulation of Bax and up-regulation of Bcl-2 and survivin, which results in alterations in apoptosis status. Differential FHIT expression in human CRC plays a role in control of cell growth and apoptosis. FHIT is a potentially important growth suppressor gene that plays a role in the development and progression of human CRC. FHIT may be a candidate for therapeutic modulation of apoptosis in human CRC.
We thank Dr. Ziqiang Yuan at Department of Molecular Genetics, Albert Einstein College of Medicine, NY, USA for revising the manuscript.
S- Editor Liu Y L- Editor Wang XL E- Editor Ma WH
| 1. | Hawk ET, Levin B. Colorectal cancer prevention. J Clin Oncol. 2005;23:378-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 155] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 2. | Li M, Gu J. Changing patterns of colorectal cancer in China over a period of 20 years. World J Gastroenterol. 2005;11:4685-4688. [PubMed] |
| 3. | Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3; 8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 736] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 4. | Boldog F, Gemmill RM, West J, Robinson M, Robinson L, Li E, Roche J, Todd S, Waggoner B, Lundstrom R. Chromosome 3p14 homozygous deletions and sequence analysis of FRA3B. Hum Mol Genet. 1997;6:193-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 92] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Hayashi S, Tanimoto K, Hajiro-Nakanishi K, Tsuchiya E, Kurosumi M, Higashi Y, Imai K, Suga K, Nakachi K. Abnormal FHIT transcripts in human breast carcinomas: a clinicopathological and epidemiological analysis of 61 Japanese cases. Cancer Res. 1997;57:1981-1985. [PubMed] |
| 6. | Huebner K, Hadaczek P, Siprashvili Z, Druck T, Croce CM. The FHIT gene, a multiple tumor suppressor gene encompassing the carcinogen sensitive chromosome fragile site, FRA3B. Biochim Biophys Acta. 1997;1332:M65-M70. [PubMed] |
| 7. | Siprashvili Z, Sozzi G, Barnes LD, McCue P, Robinson AK, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci USA. 1997;94:13771-13776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 265] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 8. | Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, Fondon JW, Garner HR, McKay B, Latif F. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000;60:1949-1960. [PubMed] |
| 9. | Sozzi G, Sard L, De Gregorio L, Marchetti A, Musso K, Buttitta F, Tornielli S, Pellegrini S, Veronese ML, Manenti G. Association between cigarette smoking and FHIT gene alterations in lung cancer. Cancer Res. 1997;57:2121-2123. [PubMed] |
| 10. | Campiglio M, Pekarsky Y, Menard S, Tagliabue E, Pilotti S, Croce CM. FHIT loss of function in human primary breast cancer correlates with advanced stage of the disease. Cancer Res. 1999;59:3866-3869. [PubMed] |
| 11. | Pavelić K, Krizanac S, Cacev T, Hadzija MP, Radosević S, Crnić I, Levanat S, Kapitanović S. Aberration of FHIT gene is associated with increased tumor proliferation and decreased apoptosis-clinical evidence in lung and head and neck carcinomas. Mol Med. 2001;7:442-453. [PubMed] |
| 12. | Hibi K, Taguchi M, Nakamura H, Hirai A, Fujikake Y, Matsui T, Kasai Y, Akiyama S, Ito K, Takagi H. Alternative splicing of the FHIT gene in colorectal cancers. Jpn J Cancer Res. 1997;88:385-388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Luceri C, Guglielmi F, De Filippo C, Caderni G, Mini E, Biggeri A, Napoli C, Tonelli F, Cianchi F, Dolara P. Clinicopathologic features and FHIT gene expression in sporadic colorectal adenocarcinomas. Scand J Gastroenterol. 2000;35:637-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Hao XP, Willis JE, Pretlow TG, Rao JS, MacLennan GT, Talbot IC, Pretlow TP. Loss of fragile histidine triad expression in colorectal carcinomas and premalignant lesions. Cancer Res. 2000;60:18-21. [PubMed] |
| 15. | Thiagalingam S, Lisitsyn NA, Hamaguchi M, Wigler MH, Willson JK, Markowitz SD, Leach FS, Kinzler KW, Vogelstein B. Evaluation of the FHIT gene in colorectal cancers. Cancer Res. 1996;56:2936-2939. [PubMed] |
| 16. | Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2991] [Cited by in RCA: 2977] [Article Influence: 110.3] [Reference Citation Analysis (0)] |
| 17. | Knösel T, Emde A, Schlüns K, Chen Y, Jürchott K, Krause M, Dietel M, Petersen I. Immunoprofiles of 11 biomarkers using tissue microarrays identify prognostic subgroups in colorectal cancer. Neoplasia. 2005;7:741-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Cao J, Li WL, Du H, Tang WB, Wang H. FHIT and p53 expression in human colonic carcinoma. Chin J Exp Surg. 2006;23:787-789. |
| 19. | Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1121] [Cited by in RCA: 1130] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 20. | Druck T, Hadaczek P, Fu TB, Ohta M, Siprashvili Z, Baffa R, Negrini M, Kastury K, Veronese ML, Rosen D. Structure and expression of the human FHIT gene in normal and tumor cells. Cancer Res. 1997;57:504-512. [PubMed] |
| 21. | Virgilio L, Shuster M, Gollin SM, Veronese ML, Ohta M, Huebner K, Croce CM. FHIT gene alterations in head and neck squamous cell carcinomas. Proc Natl Acad Sci USA. 1996;93:9770-9775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 169] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1532] [Cited by in RCA: 1574] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 23. | Reed JC. Bcl-2 and the regulation of programmed cell death. J Cell Biol. 1994;124:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1619] [Cited by in RCA: 1686] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 24. | Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581-8589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 686] [Article Influence: 31.2] [Reference Citation Analysis (0)] |