Published online Dec 28, 2007. doi: 10.3748/wjg.v13.i48.6568
Revised: July 14, 2007
Accepted: August 26, 2007
Published online: December 28, 2007
AIM: To identify and understand the regular distribution pattern and primary penetration site for Salmonella enteritidis (S. enteritidis) in the gastrointestinal tract.
METHODS: Based on the species-specific DNA sequence of S. enteritidis from GenBank, a species-specific real-time, fluorescence-based quantitative polymerase chain reaction (FQ-PCR) was developed for the detection of S. enteritidis. We used this assay to detect genomic DNA of S. enteritidis in the gastrointestinal tract, including duodenum, jejunum, ileum, cecum, colon, rectum, esophagus and stomach, from mice after oral infection.
RESULTS: S. enteritidis was consistently detected in all segments of the gastrointestinal tract. The jejunum and ileum were positive at 8 h post inoculation, and the final organ to show a positive result was the stomach at 18 h post inoculation. The copy number of S. enteritidis DNA in each tissue reached a peak at 24-36 h post inoculation, with the jejunum, ileum and cecum containing high concentrations of S. enteritidis, whereas the duodenum, colon, rectum, stomach and esophagus had low concentrations. S. enteritidis began to decrease and vanished at 2 d post inoculation, but it was still present up to 5 d post inoculation in the jejunum, ileum and cecum, without causing apparent symptoms. By 5 d post inoculation, the cecum had significantly higher numbers of S. enteritidis than any of the other areas (P < 0.01), and this appeared to reflect its function as a repository for S. enteritidis.
CONCLUSION: The results provided significant data for clarifying the pathogenic mechanism of S. enteritidis in the gastrointestinal tract, and showed that the jejunum, ileum and cecum are the primary sites of invasion in normal mice after oral infection. This study will help to further understanding of the mechanisms of action of S. enteritidis.
-
Citation: Deng SX, Cheng AC, Wang MS, Cao P. Gastrointestinal tract distribution of
Salmonella enteritidis in orally infected mice with a species-specific fluorescent quantitative polymerase chain reaction. World J Gastroenterol 2007; 13(48): 6568-6574 - URL: https://www.wjgnet.com/1007-9327/full/v13/i48/6568.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i48.6568
Salmonella is an enteric pathogen that colonizes the intestinal tract of a variety of animals, especially humans and poultry, and accounts for millions of cases of gastroenteritis and food-borne illness each year[1,2]. Salmonella enteritidis (S. enteritidis) can be transmitted to humans through the food production chain, and undercooked or raw eggs and poultry meat are a particularly high risk for humans[3]. In the last few decades, S. enteritidis has emerged as a major cause of food-borne illness worldwide. As a result of the increased prevalence of S. enteritidis and its complex life cycle, identifying the regular distribution pattern of S. enteritidis in the gastrointestinal tract will help to understand its mechanism of action.
Previous studies have shown that orally introduced S. enteritidis has a rapid transit time through the intestine, and a small proportion of the inoculum establishes itself within the walls of the small intestine and cecum several days before systemic infection[4,5]. However, the primary site of infection and the route taken by the organism to reach the heart, liver and spleen, are unclear. Infection with Salmonella is usually started by oral ingestion of the pathogen, and is followed by bacterial colonization of the gut and invasion of internal tissues. Therefore, knowledge about the mechanisms leading to invasive infections may ultimately lead to new insights into prevention and therapy.
We evaluated the specific fragment (SdfI) reported by Agron[6], which was screened for using the Supression Subtractive Hybridization method. SdfIappears to only be found in serovar enteritidis strains, which includes a wide range of clinical and environmental species. Here, based on the specific DNA sequence of serovar enteritidis, a specific fluorescent quantitative polymerase chain reaction (FQ-PCR) for detection of serovar enteritidis was developed and applied to the study of the gastrointestinal tract distribution of S. enteritidis.
A total of 14 Salmonella strains were included in this study. Most strains were purchased from the National Center for Medical Culture Collection, including S. enteritidis (Human, No. 50041), S. enteritidis (Human, No. 50040), S. enteritidis (Mouse, No. 50338), Salmonella Choleraesuis (No. 50191-1), Salmonella Typhi (No. 50013), Salmonella Typhimurium (No. 50115-13), Salmonella Paratyphi (No. 50001-24), Salmonella Pullorum (No. 50047-2), Salmonella Anatum (No. 50083-4), Salmonella Gallinarum (No. 50770), Salmonella Dublin (No. 50761). Three strains were isolated and maintained by the Research Center of Poultry Diseases, Collage of Animal Science and Veterinary Medicine of Sichuan Agricultural University, including S. enteritidis (Duck, No. MY1), S. enteritidis (Duck, No. SC1) and S. enteritidis (Chicken, No. CD1).
For the bacterial samples, 5 mL of an overnight culture grown in Luria-Bertani broth was prepared, and then the S. enteritidis cells were harvested by centrifugation. The pellet was resuspended in lysozyme solution, followed by lysis using 10% SDS at 60°C for 1 h. DNA was purified by extraction with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1). Then, a 1/10 volume of 3 mol/L sodium acetate and 2 vols absolute ethanol were added, and the nucleic acid was then pelleted by centrifugation, washed with 70% ethanol, and dried under vacuum. The DNA genomic pellet was resuspended in 40 μL TE buffer, and stored at -20°C until use.
A conventional PCR was carried out using a template from S. enteritidis (Mouse, No. 50338), with primers F1 and R1 (designed with SdfI, GenBank Accession No. AF370707.1, generated by TakaRa Biotech, Dalian, China). The primer sequences from 5' to 3' were as follows: F1, TGTGTTTTATCTGATGCAAGAGG; and R1, CGTTCTTCTGGTACTTACGATGAC. Amplification was carried out in a total volume of 50 μL, containing 1 μL each primer (25 μmol/L), 1 μL dNTPs (10 mmol/L), 2.5 U Taq DNA Polymerase (TaKaRa Taq; TakaRa Biotech), 5 μL 10 × PCR buffer (with Mg2+, 25 mmol/L) and 2 μL templates, which were then made up to a volume of 50 μL with deionized water. An initial denaturation at 95°C for 5 min was followed by 32 cycles of denaturation at 94°C for 30 s, annealing at 52.5°C for 30 s, and extension at 72°C for 40 s. Finally, an additional extension was achieved for 10 min at 72°C. The product size was 293 bp.
Finally, the product was gel-excised and quantified with appropriate standards. Its concentration was determined spectrophotometrically using the Bio-Rad Smartspec-3000, according to the manufacturer's instructions. The standards were diluted, divided into aliquots, and frozen before use.
The FQ-PCR assay, including volume, Mg2+ concentration, probe and primer concentrations, and annealing temperature were optimized. Subsequently, the sensitivity of the assay, the linear range and standard curve were determined by using known amounts of purified template DNA (generated as described above). The primers (F2 and R2) and TaqMan-probe (FP) of FQ-PCR were designed using an internal region of the 293 bp sequences (described above, generated by TakaRa Biotech), and were used as follows, from 5' to 3': F2, TTGATGTGGTTGGTTCGTCACT; R2, TCCCTGAATCTGAGAAAGAAAAACTC; and TaqMan-probe (FP), FAM-TGCAGCGAGCATGTTCTGGAAAGC-TAMRA. Amplification was carried out in a total volume of 25 μL, containing 0.6 μL each primer (10 μmol/L), 0.75 μL dNTPs (10 mmol/L), 1.25 U Ex Taq DNA Polymerase (TaKaRa Ex Taq Hot Start Version; TakaRa Biotech), 5 × PCR buffer (free Mg2+) 5 μL, 0.8 μL TaqMan probe (5 μmol/L), 0.5 μL Mg2+ (250 mmol/L) and 1 μL templates, then made up to a volume of 25 μL with deionized water. Each run consisted of a 95°C 5 min hot start, which activated the conjugated polymerase, followed by 45 cycles (50 cycles for sensitivity experiments) with 94°C denaturation for 30 s, 55°C annealing for 30 s, and reading the fluorescent signal at this step.
The primers of FQ-PCR (F2 and R2) were used for conventional PCR with S. enteritidis (Mouse, No. 50338) DNA templates, in order to verify the specific amplification. Amplification was carried out in a total volume of 50 μL, containing 1 μL each primer (25 μmol/L), 1 μL dNTPs (10 mmol/L), 2.5 U Taq DNA polymerase (TaKaRa Taq; TakaRa Biotech), 10 × PCR buffer (with Mg2+, 25 mmol/L) 5 μL, and 2 μL templates, then made up to a volume of 50 μL with deionized water. Initial denaturation at 95°C for 5 min was followed by 32 cycles of denaturation at 94°C for 30 s, annealing at 49°C for 30 s, and extension at 72°C for 40 s. Finally, an additional extension was achieved for 10 min at 72°C. A 10 μL aliquot of PCR product was electrophoresed on a 1.5% agarose gel for 40-50 min at 80 V, and visualized and photographed under UV illumination.
Based on previous studies, the standard curve was generated as follows[7-9]: Standard DNA from S. enteritidis was used to establish a standard curve. The standard DNA contained amplified target DNA in different quantities which was included in each LightCycler run. The Primers F2 and R2 were used for this amplification, and a range of 9 × 108 - 9 × 103 gene copies was used. Concentrations of the standards were measured by fluorometric analysis, and an analysis of key LightCycler measures was performed after each run to verify identical amplification efficiencies and conditions between runs. Finally, these data were used to generate the standard curve through the iCycler IQ Detection System (Bio-Rad, USA) software.
To evaluate the variability between experiments, four different known concentrations of DNA were amplified by performing the assay described above in triplicate. For each experiment, the crossing point, average crossing point, standard deviation, and coefficient of variation for each assay were calculated.
To determine the detection limit of this FQ-PCR assay, different quantities of standard DNA and different cell numbers of S. enteritidis were introduced in the FQ-PCR. A range of 4 × 103 - 4 × 10-1 copies of standard DNA of S. enteritidis was added. Then, the results of each concentration were measured by fluorometric analysis. Bacterial cell limit detection was as follows: S. enteritidis (Mouse, No. 50338) grown aerobically at 37°C for 18 h in 5 mL Luria-Bertani broth. Subsequently, 1 mL of overnight culture was harvested, and the pellet was resuspended in 500 μL TE buffer (pH 8.0), from which the number of S. enteritidis cells was obtained by conventional viable count method. Then, a 10 fold serial dilution was performed on the overnight culture liquor, and the phenol/chloroform/isoamyl alcohol method was used to extract the DNA template serially from 10-6 - 10-10 serial dilutions of bacterial liquor. Finally, 1 μL aliquot per concentration of the DNA template was subjected to a test of bacterial cell sensitivity, and the result of the viable count was used to obtain the final result. The FQ-PCR was performed and analyzed as described above.
All 14 bacterial strains were used to assess the specificity of the FQ-PCR. The boiling method was used to prepare the DNA template, and 4 μL of this was used in FQ-PCR.
Our infection model was based on previous studies, which showed that orally introduced S. enteritidis had a rapid transit time through the intestine and establishes itself within the walls of the gut for more than 3 d[4,5,10-12,20]. Twenty-eight mice (age 9 wk, specific-pathogen-free) were purchased from the Animal Center of Sichuan University, China. In brief, a group of 14 white mice were orally infected with a virulent S. enteritidis strain (Mouse, No. 50338), at 2.0 × 104 cells per mouse. Another group of 14 white mice was treated with an equal volume of water as a control. Duodenum, jejunum, ileum, cecum, colon, rectum, esophagus and stomach were analyzed by FQ-PCR at different post-inoculation time points, at 30 min, 1, 2, 4, 8, 12, 16, 18, 24 and 36 h, and 2, 3, 4 and 5 d.
One mouse from each group was sacrificed at each time point and its organs were aseptically removed and immediately placed in 1.5 mL labeled snap-cap tubes. The contents were gently removed by lightly squeezing the excised organ and washing twice with 0.85% NaCl. Finally, the tissue samples, without contents in lumens, were placed in 1.5 mL labeled snap-cap tubes and frozen before use.
DNA extraction from the tissue samples was as follows. 0.3 g tissue samples from different segments of the gastrointestinal tract were washed with 0.85% NaCl twice in order to confirm the removal of contents in lumens. Then, the samples were ground in 1.5 mL Eppendorf tubes using a conventional method. The pellet was resuspended in 500 μL TE buffer (pH 8.0) with 10 μL proteinase K (30 mg/mL) and incubated at 37°C for 3 h. Finally, a conventional phenol/chloroform/isoamyl alcohol method (preparation described above) was used to extract the genomic DNA of S. enteritidis from tissue, and 1 μL aliquot of the sample DNA template was used for FQ-PCR detection.
The primers of FQ-PCR were used for conventional PCR with S. enteritidis (Mouse, No. 50338) DNA templates, in order to verify the specific amplification. Results showed that the PCR produced an intense band with the expected 130 bp for S. enteritidis, which indicated 100% specificity.
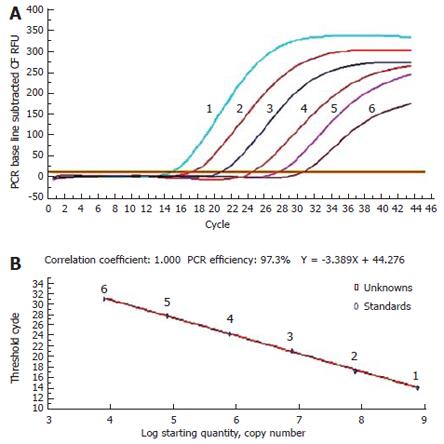
One of the main advantages of FQ-PCR is the ability to quantitate unknown samples. With this assay, it is possible to carry out a rapid quantitative analysis of DNA over a wide linear range, with an unknown template. By using a standard template containing from 9 × 108 - 9 × 103 copies, accurate results for a series of samples were obtained, based on the data used to generate the standard curve with the iCycler IQ Detection System. The correlation coefficient for the associated standard curve was 1.000 and PCR efficiency was 97.3%, indicating that the crossing threshold values for the standards fell within an acceptable range. Using the following formula, we could quantitate the number of DNA copies of S. enteritidis for unknown samples: Y= -3.389X + 44.276 (where Y is the threshold cycle, and X the log of the starting quantity) (Figure 1).
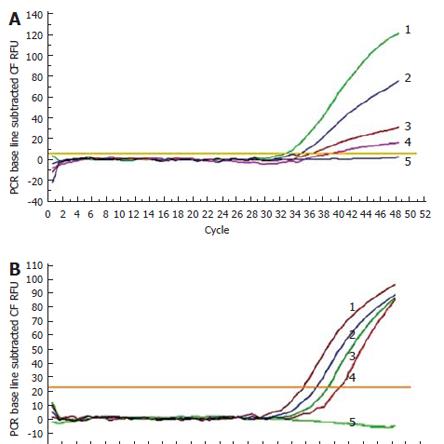
A range of 4.0 × 103-4.0 × 10-1 copies of the S. enteritidis standard template was used, and the limit of detection was 4 copies/μL. A sensitivity of 6 cfu/mL was obtained when 10-fold serial dilutions of bacterial cell cultures were used as the PCR template (Figure 2).
Four different, known concentrations of DNA (1.2 × 109-1.2 × 106 copies/μL) were amplified by performing the assay described above in triplicate. Analysis of these values proved that the assay was reproducible, as the coefficient of variation was statistically low, at < 1.5%. The threshold cycle for each concentration ranged from 1.2 × 109-1.2 × 106 copies/μL and was different between 0.1-0.3 cycles and highly reproducible (Figure 3).
All 14 bacterial strains used to assess the specificity of the PCR indicated that only S. enteritidis genomic strains showed positive results, while there was no amplification with non-S. enteritidis strains (data not shown).
The distribution of S. enteritidis within the gastrointestinal tract after oral infection was determined by means of FQ-PCR at intervals on separate segments of gut over a 5-d period. Results showed that the jejunum and ileum were positive at 8 h postinoculation, with approximately 200 copies/g. Then, S. enteritidis was consistently detected in all segments of the gastrointestine at 8 h post inoculation, and the last organ to show a positive result was the stomach at 18 h post inoculation. The copy numbers of S. enteritidis in each tissue reached a peak at 24-36 h, with the jejunum, ileum and cecum containing high concentrations of S. enteritidis, whereas the duodenum, colon, rectum, esophagus and stomach had low concentrations. Numbers of bacteria decreased at 2-3 d, and by 4 d the level of S. enteritidis had clearly decreased, with the duodenum, colon, esophagus and stomach not having a positive result. The rectum had about 10 copies/g at 4 d post inoculation, and then the bacteria vanished. The rectum carried the S. enteritidis for up to 5 d in others without causing apparent symptoms. Importantly, with respect to the number of S. enteritidis cells at 5 d postinoculation, compared to the other organs the cecum had significantly higher numbers of S. enteritidis than all of the other regions, with 103.3 copies/g, whereas the jejunum had 101.5 copies/g, and the ileum had 101.3 copies/g (P < 0.01). The control group did not have any positive results at any time in any location. The details are given in Table 1.
| Time | DuodeNum | Jejunum | Ileum | Cecum | Colon | Rectum | Esophagus | Stomach |
| 30 min | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 h | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 h | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 h | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 8 h | 0.0 | 2.3 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 h | 1.2 | 2.5 | 2.8 | 2.1 | 1.3 | 1.6 | 0.0 | 0.0 |
| 16 h | 1.5 | 3.3 | 3.2 | 2.6 | 1.3 | 1.6 | 1.1 | 0.0 |
| 18 h | 2.2 | 3.3 | 3.6 | 3.0 | 2.2 | 1.8 | 1.2 | 1.0 |
| 24 h | 3.0 | 4.0 | 4.2 | 3.9 | 2.6 | 2.1 | 1.4 | 1.2 |
| 36 h | 3.3 | 6.2 | 6.6 | 5.4 | 2.6 | 3.5 | 2.6 | 1.9 |
| 2 d | 2.2 | 4.1 | 4.0 | 3.9 | 1.6 | 2.6 | 1.6 | 1.7 |
| 3 d | 1.2 | 3.0 | 2.8 | 3.6 | 1.2 | 1.2 | 1.3 | 1.2 |
| 4 d | 0.0 | 2.5 | 2.3 | 3.4 | 0.0 | 1.1 | 0.0 | 0.0 |
| 5 d | 0.0 | 1.5 | 1.3 | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 |
Attachment to host tissues is the first important step for establishing a bacterial infection[13,14]. The fimbria Sef 14 is found in a limited number of Salmonella enterica serovars, including enteritidis. This surface structure appears to be required for macrophage uptake and survival in intraperitoneal infections[15], in contrast to other Salmonella fimbriae that promote binding to host epithelial cells[16]. There is also evidence that quorum sensing plays an important role in the life cycle of Salmonella serovar enteritidis[17]. Upon interaction with the intestinal epithelium, Salmonella can induce changes in the brush border known as membrane ruffles, survive and multiply there[18]. This study showed that S. enteritidis in orally infected mice very quickly reached the intestinal tract, especially the jejunum and ileum. We suggest that penetration by S. enteritidis occurs in the upper half of the gastrointestinal tract, which possibly indicates sites that are more easily penetrated by S. enteritidis. This finding is compatible with results obtained in previous studies, which showed that S. enteritidis has a rapid transit time through the normal mouse intestine; a small proportion of the inoculum establishes itself within the walls of the small intestine, before a systemic infection can be demonstrated[4,5,19]. However, these studies were unable to precisely establish the primary site of bacterial invasion in these animals. From our study, it seems reasonable that the jejunum and ileum are most likely sites for bacterial penetration that is responsible for systemic infection. First, the jejunum and ileum were positive at 8 h post inoculation. Second, the S. enteritidis copy number reached a maximum between 8 h and 36 h post inoculation, with copies of S. enteritidis being about 10-10 000 times that of other regions. Finally, by 4 d post inoculation, the level of S. enteritidis clearly decreased, with no bacteria in the duodenum, colon, esophagus and stomach, but S. enteritidis remained in the jejunum, ileum and cecum for up to 5 d post inoculation. At 12 h post inoculation, the S. enteritidis in the cecum were increasingly obvious, but less so in the jejunm and ileum (about 10 times fewer) at 24-36 h post inoculation. By 5 d post inoculation, the cecum had significantly higher numbers of S. enteritidis than all the other areas, with 103.3 copies/g, whereas the jejunum had 101.5 copies/g, and the ileum had 101.3 copies/g (P < 0.01). Therefore, this appeared to reflect the function of the cecum as a repository for S. enteritidis and a site for its penetration. By way of contrast, Ozawa[20] has concluded that the primary site of Salmonella infection involves the cecum and the large intestine.
Mucosal immunity provides the first line of protection following oral exposure to pathogens. In particular, the involvement of mucosal IgA in protection against salmonellosis has been reported[21]. Secretory IgA limits mucosal colonization by S. enteritidis by preventing adherence and subsequent invasion of the bacteria[22,23]. The environment of the gastrointestinal tract can also prevent invasion by S. enteritidis to some extent. The lowest concentration of S. enteritidis was found in the stomach, and this was the last area in which S. enteritidis was detected, which indicates that environmental factors, such as pH, can influence the survival and virulence of S. enteritidis. It has been reported that normal host defenses are capable of eliminating > 80% of organisms from the gut within hours[20]. Most of the Salmonella in the challenge inoculum had no pathogenic significance, since only a few organisms passed through the mucosa of the ileum during the initial hours of infection. A previous study has shown that S. enteritidis colonization in the gastrointestinal tract can persist for as long as 18 wk post inoculation in hens[24]. Nevertheless, these few organisms have the potential to cause a lethal systemic infection[20]. Changes in the microenvironment in the gastrointestinal tract have important implications for understanding the gastrointestinal factors necessary for protection against S. enteritidis infection. S. enteritidis is distributed evenly along the intestinal tract, and the microenvironment may be a predictor for severity of S. enteritidis invasion and infection[25,26].
FQ-PCR has become a potentially powerful alternative in microbiological diagnosis due to its simplicity, rapidity, reproducibility and accuracy. The specific primer-probe combination is a powerful tool for detecting the genetic content of closely related bacterial species[27,28]. A series of sensitivity experiments was performed and proved that the detection limit of this method was 4 copies/μL for standards template, and 6 cfu/mL for bacterial cell number. Four different, known concentrations of DNA were amplified by performing the assay described above in triplicate, and showed a coefficient of variation less than 1.5%. Also, one of the main advantages of FQ-PCR is the ability to quantitate unknown samples. With this assay, it is possible to carry out a rapid quantitative analysis of DNA over a wide linear range, with an unknown template. Simultaneously, variation in results may be due to the extraction efficiency of DNA, the PCR inhibitors, or a large amount of DNA from background organisms.
In conclusion, our results provide significant data for clarifying the pathogenic mechanism of S. enteritidis in the gastrointestinal tract, and show that the jejunum, ileum and cecum are the primary sites of invasion in normal mice after oral infection. This study will help to understand the mechanisms of action of S. enteritidis.
There are over 2500 serovars in the genus Salmonella. It has been a public health concern for over 100 years, and the incidence of Salmonella infections has risen dramatically, especially those caused by S. enteritidis. Therefore, knowledge about Salmonella infection could be an additional means for decreasing the incidence of infection. Infection with Salmonella is usually started by oral ingestion of the pathogen, and is followed by bacterial colonization of the gut and invasion of internal tissues. Therefore, it is necessary to understand its mechanisms of action in the gastrointestinal tract.
To date, the exact site of primary penetration of S. enteritidis in the gut has not been established. FQ-PCR, as a rapid, sensitive technique for precise quantitation of nucleic acid, will be an ideal method to study the distribution of S. enteritidis in the gastrointestinal tract.
Previous studies have shown that S. enteritidis establishes itself within the walls of the small intestine before a systemic infection can be demonstrated. However, these studies have been unable to identify the regular distribution pattern and the primary site of S. enteritidis invasion. In this study, we offered a significant improvement over previous studies, and suggested that the jejunum, ileum and cecum are the primary sites of invasion after oral infection.
This study will provide significant data for clarifying the pathogenic mechanisms of S. enteritidis in the gastrointestinal tract, and may ultimately lead to new insights in prevention and therapy.
Standard DNA template: The purified target DNA fragment for FQ-PCR amplification that is usually used to generate standard curves. It can be generated by gel excised directly after a conventional PCR amplification or after the target fragment is cloned using plasmid DNA.
This study detected FQ-PCR S. enteritidis in different segments of the gastrointestinal tract in mice after oral infection, and demonstrated the regular distribution pattern and primary penetration sites of S. enteritidis. The method is simple and accurate, and it may lead to new ways to prevent S. enteritidis infection.
S- Editor Liu Y L- Editor Kerr C E- Editor Ma WH
| 1. | Palmer S, Parry S, Perry D, Smith R, Evans M, Nehaul L, Roberts R, Walapu M, Wright D. The role of outbreaks in developing food safety policy: population based surveillance of salmonella outbreaks in Wales 1986-98. Epidemiol Infect. 2000;125:467-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Van Loock F, Ducoffre G, Dumont JM, Libotte-Chasseur ML, Imberechts H, Gouffaux M, Houins-Roulet J, Lamsens G, De Schrijver K, Bin N. Analysis of foodborne disease in Belgium in 1997. Acta Clin Belg. 2000;55:300-306. [PubMed] |
| 3. | Gillespie IA, O'Brien SJ, Adak GK, Ward LR, Smith HR. Foodborne general outbreaks of Salmonella Enteritidis phage type 4 infection, England and Wales, 1992-2002: where are the risks. Epidemiol Infect. 2005;133:795-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | Collins FM. Immunity to enteric infection in mice. Infect Immun. 1970;1:243-250. [PubMed] |
| 5. | Collins FM. Salmonellosis in orally infected specific pathogen-free C57B1 mice. Infect Immun. 1972;5:191-198. [PubMed] |
| 6. | Agron PG, Walker RL, Kinde H, Sawyer SJ, Hayes DC, Wollard J, Andersen GL. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl Environ Microbiol. 2001;67:4984-4991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 121] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Jothikumar N, Cromeans TL, Hill VR, Lu X, Sobsey MD, Erdman DD. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl Environ Microbiol. 2005;71:3131-3136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 193] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 8. | Skovhus TL, Ramsing NB, Holmström C, Kjelleberg S, Dahllöf I. Real-time quantitative PCR for assessment of abundance of Pseudoalteromonas species in marine samples. Appl Environ Microbiol. 2004;70:2373-2382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Kasai M, Francesconi A, Petraitiene R, Petraitis V, Kelaher AM, Kim HS, Meletiadis J, Sein T, Bacher J, Walsh TJ. Use of quantitative real-time PCR to study the kinetics of extracellular DNA released from Candida albicans, with implications for diagnosis of invasive Candidiasis. J Clin Microbiol. 2006;44:143-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Desmidt M, Ducatelle R, Haesebrouck F. Research notes: Immunohistochemical observations in the ceca of chickens infected with Salmonella enteritidis phage type four. Poult Sci. 1998;77:73-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Hohmann AW, Schmidt G, Rowley D. Intestinal colonization and virulence of Salmonella in mice. Infect Immun. 1978;22:763-770. [PubMed] |
| 12. | Takata T, Liang J, Nakano H, Yoshimura Y. Invasion of Salmonella enteritidis in the tissues of reproductive organs in laying Japanese quail: an immunocytochemical study. Poult Sci. 2003;82:1170-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Turnbull PC, Richmond JE. A model of salmonella enteritis: the behaviour of Salmonella enteritidis in chick intestine studies by light and electron microscopy. Br J Exp Pathol. 1978;59:64-75. [PubMed] |
| 14. | Amin II, Douce GR, Osborne MP, Stephen J. Quantitative studies of invasion of rabbit ileal mucosa by Salmonella typhimurium strains which differ in virulence in a model of gastroenteritis. Infect Immun. 1994;62:569-578. [PubMed] |
| 15. | Edwards RA, Schifferli DM, Maloy SR. A role for Salmonella fimbriae in intraperitoneal infections. Proc Natl Acad Sci USA. 2000;97:1258-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Dibb-Fuller MP, Allen-Vercoe E, Thorns CJ, Woodward MJ. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology. 1999;145:1023-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 123] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Guard-Petter J. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl Environ Microbiol. 1998;64:2166-2172. [PubMed] |
| 18. | Takeuchi A. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967;50:109-136. [PubMed] |
| 19. | Sprinz H, Gangarosa EJ, Williams M, Hornick RB, Woodward TE. Histopathology of the upper small intestines in typhoid fever. Biopsy study of experimental disease in man. Am J Dig Dis. 1966;11:615-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 49] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Carter PB, Collins FM. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 496] [Cited by in RCA: 473] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 21. | Hassan JO, Porter SB, Curtiss R 3rd. Effect of infective dose on humoral immune responses and colonization in chickens experimentally infected with Salmonella typhimurium. Avian Dis. 1993;37:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904-3913. [PubMed] |
| 23. | Sheela RR, Babu U, Mu J, Elankumaran S, Bautista DA, Raybourne RB, Heckert RA, Song W. Immune responses against Salmonella enterica serovar enteritidis infection in virally immunosuppressed chickens. Clin Diagn Lab Immunol. 2003;10:670-679. [PubMed] |
| 24. | Gast RK, Benson ST. The comparative virulence for chicks of Salmonella enteritidis phage type 4 isolates and isolates of phage types commonly found in poultry in the United States. Avian Dis. 1995;39:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Holt PS, Porter RE Jr. Microbiological and histopathological effects of an induced-molt fasting procedure on a Salmonella enteritidis infection in chickens. Avian Dis. 1992;36:610-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 62] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Holt PS, Macri NP, Porter RE Jr. Microbiological analysis of the early Salmonella enteritidis infection in molted and unmolted hens. Avian Dis. 1995;39:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L. Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl Environ Microbiol. 2003;69:3456-3461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 166] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 28. | Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 987] [Article Influence: 32.9] [Reference Citation Analysis (0)] |