Published online Dec 21, 2007. doi: 10.3748/wjg.v13.i47.6425
Revised: September 19, 2007
Accepted: November 19, 2007
Published online: December 21, 2007
AIM: To study the effectiveness and mechanisms of anti- human vascular endothelial growth factor (hVEGF) hairpin ribozyme on angiogenesis, oncogenicity and tumor growth in a hepatocarcinoma cell line and a xenografted model.
METHODS: The artificial anti-hVEGF hairpin ribozyme was transfected into hepatocarcinoma cell line SMMC-7721 and, subsequently, polymerase chain reaction (PCR) and reverse transcription polymerase chain reaction (RT-PCR) were performed to confirm the ribozyme gene integration and transcription. To determine the effects of ribozyme ,VEGF expression was detected by semiquantitative RT-PCR and enzyme liked immunosorbent assay (ELISA). MTT assay was carried out to measure the cell proliferation. Furthermore,the transfected and control cells were inoculated into nude mice respectively, the growth of cells in nude mice and angiogenesis were observed.
RESULTS: VEGF expression was down-regulated sharply by ribozyme in transfected SMMC-7721 cells and xenografted tumor. Compared to the control group, the transfected cells grew slower in cell cultures and xenografts, and the xenograft formation was delayed as well. In addition, the microvessel density of the xenografted tumor was obviously declined in the transfected group. As demonstrated by microscopy,reduction of VEGF production induced by ribozyme resulted in a significantly higher cell differentiation and less proliferation vigor in xenografted tumor.
CONCLUSION: Anti-hVEGF hairpin ribozyme can effectively inhibit VEGF expression and growth of hepatocarcinoma in vitro and in vivo. VEGF is functionally related to cell proliferation, differentiation and tumori-genesis in hepatocarcinoma.
- Citation: Li LH, Guo ZJ, Yan LL, Yang JC, Xie YF, Sheng WH, Huang ZH, Wang XH. Antitumor and antiangiogenic activities of anti-vascular endothelial growth factor hairpin ribozyme in human hepatocellular carcinoma cell cultures and xenografts. World J Gastroenterol 2007; 13(47): 6425-6432
- URL: https://www.wjgnet.com/1007-9327/full/v13/i47/6425.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i47.6425
Rapidly growing tumors routinely outstrip their supply of oxygen and nutrients, and induction of new blood vessels is critical to sustain neoplastic proliferation[1-4]. A number of growth factors have been identified as potential positive regulators of angiogenesis. Among them, vascular endothelial growth factor (VEGF) appears to have a central role in the angiogenic process. VEGF not only is the target of many proangiogenic factors but also regulates molecules that are implicated in endothelial proliferation. It has been suggested that VEGF may be a proximate angiogenic factor through which others act. In fact, over-expression of VEGF is the characteristic of most malignant tumors including hepatocarcinoma[5-9], which is regulated in response to hypoxia[7,10-12] and highly related to microvessel density of cancer, grade of malignance and metastasis[13-15]. It now appears that VEGF also has autocrine functions acting as a survival factor for tumor cells protecting themselves from stresses such as hypoxia, chemotherapy and radiotherapy[16]. Consequently, anti-VEGF therapies are being actively investigated as potential anti-cancer treatment modalities, either as alternatives or adjuncts to conventional chemo or radiation therapy. Clinical trials using anti-VEGF mAbs such as bevacizumab have validated the efficacy of this therapeutic approach but have also revealed adverse effects.
Hepatocellular carcinoma (HCC) is one of the most common and aggressive cancers worldwide, being the third cause for cancer-related deaths. As a malignant solid tumor, HCC is characterized by fast infiltrating growth, early metastasis, high-grade malignancy, and poor therapeutic efficacy[17]. It is a highly vascular tumor dependenting on neovascularization and a rapidly developing malignancy[18]. The VEGF level in HCC tissues is significantly higher than that in distal cancerous tissues[6]. The abnormal expression levels of VEGF in sera of HCC patients are directly correlated with the metastasis and recurrence of tumors[5,19,20]. Since HCC is insensitive to conventional chemotherapeutics and its prognosis is poor, it is important to explore an antiangiogenesis method for HCC characterized by a high vascularity. Up to now, many strategies for gene therapy have been developed. Ribozymes are a group of catalytically active nucleic acids capable of site-specific cleavage of target mRNAs, thus decreasing mRNA expression and inhibiting the function of target gene[21]. Both hairpin and hammerhead ribozymes are the most commonly used gene therapy models. In contrast to hammerhead ribozyme, the hairpin ribozyme reaction requires an environment closer to human physiology. The hairpin ribozyme uses a catalytic mechanism that does not require metals for cleavage or ligation of substrate RNA. In this regard, it is presently unique among RNA catalysts. The hairpin ribozyme has been approved for its use in gene therapy.
The role of VEGF in HCC proliferation, differentiation and tumorigenesis is unclear. The present study was to study the effectiveness and mechanisms of hairpin ribozyme targeting human VEGF (hVEGF) mRNA on exon 3 on VEGF expression, angiogenesis, oncogenicity and tumor growth in HCC cell cultures and xenografts.
Human hapatocarcinima cell line SMMC-7721 was maintained in our laboratory. Cells were incubated with RPMI 1640 medium (GIBCO BRL, Carlsbad, CA, USA) supplemented with 10% heat-inactivated calf serum (Sijiqing, Hangzhou, China), 100 U/mL penicillin G sodium and 100 μg/mL streptomycin sulfate in a humidified atmosphere containing 50 mL/L CO2 at 37°C.
The secondary structure of ribozyme was obtained by the ribozyme- aided design software, and the primers of hairpin ribozyme were synthesized (Shengong, Shanghai, China) based on the third exon of hVEGF as previously described[22]. The sequences of specific primers of ribozyme used are as follows: forward: 5'-GATCCCTGATAAGAATCCAACCAGAGAAACACACGTTGTGGTATATTACCTGGTAG-3' and reverse: 5'-AATTCTACCAGGTAATATACCACAACGTGTGTTTCTCTGGTTGGATTCTTATCAGG-3'. Ribozyme was synthesized in a 50 μL reaction system, which was mixed with 1 μL (25 μmol/L) primers, 5 μL NaCl buffer and 43 μL ddH2O. Since the gene of anti- hVEGF hairpin ribozyme only has a 50 bp product, after incubation at 94°C for 2 min followed by annealing of the two primers at room temperature, ribozyme was acquired.
After plasmid pcDNA3.1+ (Invitrogen, San Diego, CA) was digested with restriction endonucleases BamHI and EcoRI (TakaRa, Japan), the purified products of anti- hVEGF hairpin ribozyme were subcloned into them, thus, the recombinant eukaryotic expression plasmid pcDNA3.1+/ribozyme (abbreviated as pcDNA3.1+/RZ) was constructed.
SMMC-7721 cells were seeded in 6-well plates at 2 × 105 cells per well and cultured as described above. The recombinant eukaryotic expression plasmid pcDNA3.1+/RZ was transfected into SMMC-7721 cells by LipofetamineTM2000 (Invitrogen, San Diego, CA) following the manufacturer's instructions, and positive clones were selected with G418, then SMMC-7721/RZ transgenic cells were obtained. At the same time, two control groups were established: the SMMC-7721 cell control (abbreviated as SMMC-7721 cell) and the SMMC-7721 cells transfected with blank vector of pcDNA3.1+ control (abbreviated as SMMC-7721/pcDNA3.1+ cells).
Integration and transcription of the objective gene in SMMC-7721/RZ transgenic cells were identified by polymerase chain reaction (PCR) and reverse transcription polymerase chain reaction (RT-PCR). Genomic DNA was extracted from cells in 3 groups using genomic DNA (Shengong, Shanghai, China). PCR was performed to detect the integration of ribozyme gene in SMMC-7721/RZ transgenic cells. The ribozyme primers (forward: 5'-GGACTTTCCTACTTGGCAGTACATC-3', reverse: 5'-CCACAACGTGTGTTTCTCTGGTTGG-3') were used in this study. Genomic DNA template was amplified with an initial denaturation at 94°C for 2 min, then 30 cycles at 94°C for 50 s, at 58°C for 50 s and at 72°C for 45 s, followed by at 72°C for 10 min. To evaluate transcription of the ribozyme gene in SMMC-7721/RZ transgenic cells, RT-PCR was performed. Total RNA was extracted from cells using UNIQ column total RNA extraction kits (Shengong, Shanghai, China). First strand cDNA was synthesized from 2 μg of total RNA with an oligo (dT)18-primer in 20 μL of first strand reaction mix at 37°C for 1 h. Thirty cycles of PCR were done. First strand cDNA was employed as template and the sequences of specific primers of ribozyme are as follows: forward: 5'-TAG AGAACCCACTGCTTACTGGCT-3', and reverse: 5'-CCACAACGTGTGTTTCTCTGGTTGG-3'.
SMMC-7721/RZ cells were seeded in 24-well plates at 1 × 105 cells per well. The SMMC-7721 cell control and the SMMC-7721/pcDNA3.1+ cell control were established. Cells were cultured for 72 h, and then the supernatant was harvested. Total RNA was isolated from cells, and then RT-PCR was performed to detect the transcription level of VEGF mRNA in cells. Data were presented and normalized to β-actin. The sequences of PCR primers of VEGF and β-actin are as follows: VEGF (forward: 5'-TGGTAGAGTTCATGGATGTCTATCA-3', reverse: 5'-GCATGGTGATGTTGGACTCCTCA-3'), β-actin (forward: 5'-TGCGTGACATTAAGGAGAAG-3', reverse: 5'-CTGCATCCTGTCGGCAATG-3'). The expression of VEGF protein in supernatant was detected by ELISA assay following the manufacturer's instructions (Jingmei, Shanghai, China).
SMMC-7721/RZ cells were seeded in 96-well plates at 1 × 104 cells per well. Three control groups were established as follows: non-cell control group, SMMC-7721 cell control group and SMMC-7721/pcDNA3.1+ cell control group. Cells were cultured for 24, 48, 72, 96 h respectively, and viability was assessed using 3-(4, 5- dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) (SIGMA, St. Louis. USA) at a final concentration of 0.5 mg/mL. Cells were incubated for 2 h, the medium was aspirated, and the cells were dissolved in acidic isopropanol (90% isopropanol, 0.5% sodium dodecyl sulfate, 40 mmol/L HCl). Optical density was read on a microplate reader at 570 nm using the isopropanol as blank. The inhibitory rate (IR) was calculated as follows: IR (%) = (1-absorbance of the treated wells)/(absorbance of the control wells) × 100%.
Female athymic BLAB/c nude mice, 4-5 wk of age, were purchased from Shanghai Experimental Animal Company, Chinese Academy of Sciences (Shanghai, China) and housed in a pathogen-free facility. In vivo studies approved by the Fourth Affiliated Hospital of Soochow University Animal Care Committee, were conducted in accordance with the institutional and China guidelines. Subconfluent SMMC-7721/RZ transgenic cells or SMMC-7721/pcDNA3.1+ cells were suspended in PBS and injected subcutaneously into the right flanks (2 × 106/0.1 mL) of 5 nude mice in each treatment group. Tumor size was measured every other day using an external caliper. Tumor volumes (V) were determined following the equation: V = (L × W2) × 0.5, where L is the length and W is the width of tumor. The standard for tumor formation was the diameter of tumors > 0.5 cm. When the experiment was terminated at the 5th wk after tumor formation, the mice were sacrificed. The tumors were isolated, photographed, snap frozen in liquid nitrogen and stored at -80°C, or fixed in formalin for subsequent multiple assays.
Tumor tissues were formalin-fixed and paraffin-embedded. After deparaffinized and rehydrated, the slides were stained with hematoxylin and eosin (HE) in succession, and finally mounted and visualized under microscope. The number of karyokinesis and pathologic karyokinesis was calculated in each visual field, and ten visual fields were selected randomly in each section.
Immunostaining was performed on 5-μm thick sections. Serial sections of paraffin-embedded tumor tissue were dewaxed and hydrated. After antigen retrieval, its endogenous peroxidase activity was blocked with 3% hydrogen peroxide solution for 5-10 min. The sections were washed in distilled water, and soaked in pH 7.4 phosphate-buffered saline (PBS) for 5 min. Nonspecific binding was blocked by incubation with 3% normal goat serum for 10 min. The sections were incubated with primary antibody, rabbit anti-human VEGF polyclonal antibody or rat anti-murine cell determinant (CD34) monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz), at a dilution of 1:100 for 1-2 h at 37°C. Anti-CD34 was used as a pan-endothelial marker for microvessel density (MVD) analysis. After washed with PBS, the sections were incubated for 30 min at 37°C with biotinylated secondary antibody and then with horseradish peroxidase (HRP)-conjugated streptavidin for 30 min at 37°C. The color was developed with a Vector DAB substrate kit for 1 min and 30 s and counterstained with hematoxylin. All sections were analyzed by conventional light microscopy and digital photography.
MVD was evaluated according to the CD34 endothelial cell immunostaining. For the microvessel counting, positivelystained CD34 was counted in ten visual fields of each slide. Any endothelial cluster positive for CD34 (brown yellow staining) was considered a single countable microvessel.
All statistical analyses were carried out by the Statistical Package for the Social Sciences (SPSS) software, Version 10.0 for Windows. All values such as tumor volume and weight, karyokinesis and microvessel, were expressed as mean ± SD. Chi-square test was used for rate contrast. P < 0.05 was considered statistically significant.
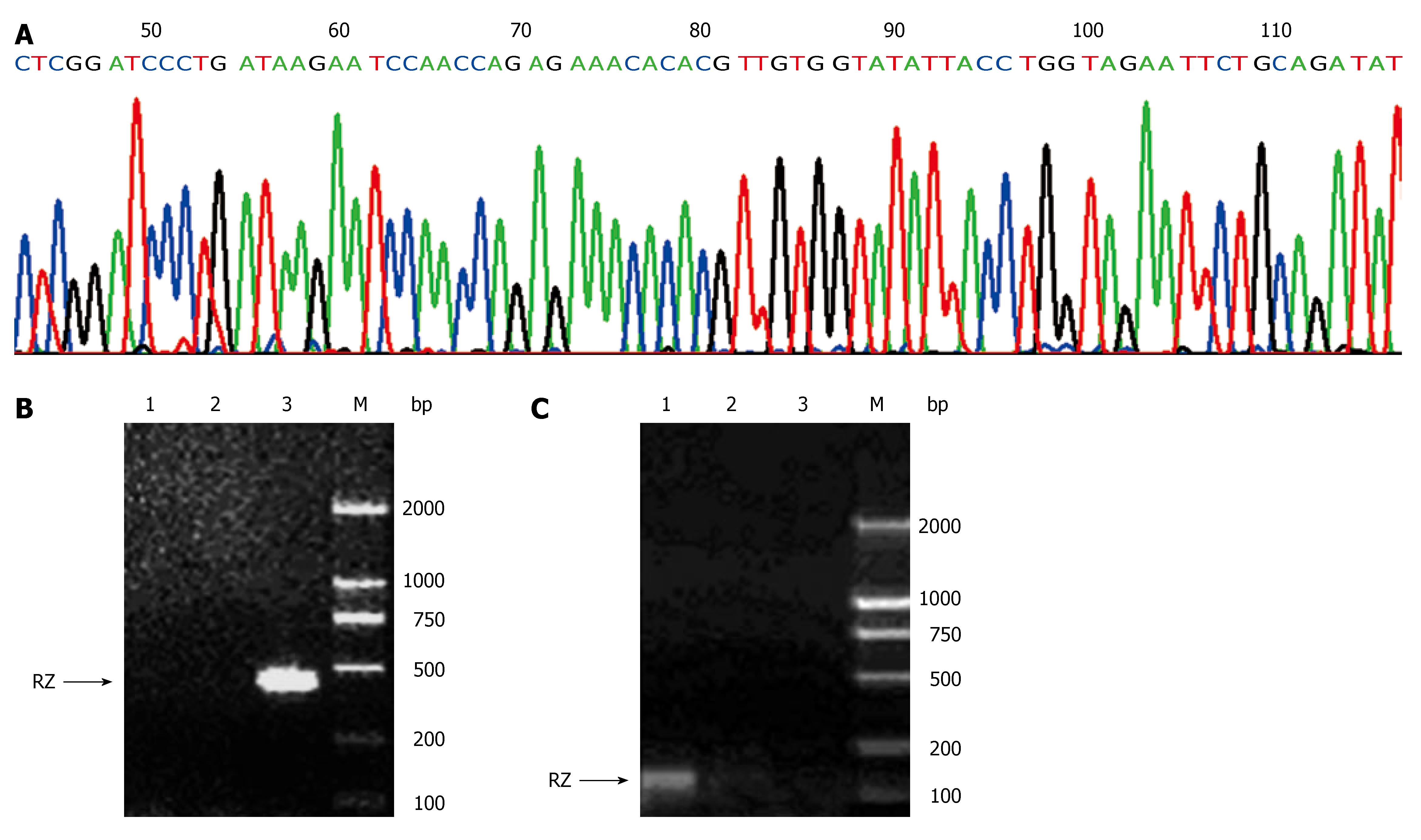
When products from annealing of the two specific primers of ribozyme were run on agarose gels and visualized with ethidium bromide staining, an amplicon of 50 bp of anti-hVEGF hairpin ribozyme (including cohesive terminal of BamHI and EcoRI) was shown. After cleaved by BamHI and EcoRI, the recombinant eukaryotic expression plasmid pcDNA3.1+/RZ generated an amplicon of 50 bp, revealing that anti-hVEGF hairpin ribozyme was successfully subcloned into pcDNA3.1+. Recombinant plasmids harboring ribozyme sequences in the pcDNA3.1+ backbone were checked by restriction enzyme analysis and sequencing. As shown in Figure 1A, the sequence of the gene subcloned into pcDNA3.1+ was identical to that of the synthetic gene designed according to Genebank, indicating that the recombinant eukaryotic expression plasmid pcDNA3.1+/RZ was successfully constructed.
The forward and reverse primers were designed based on the sequence of pcDNA3.1+ and anti-hVEGF hairpin ribozyme. PCR amplification of ribozyme in genomic DNA of SMMC-7721/RZ cells generated an amplicon of 400 bp (Figure 1B). Using the primers designed based on the sequence of pcDNA3.1+ transcriptional start site and ribozyme, RT-PCR showed that PCR amplification of ribozyme in cDNA of SMMC-7721/RZ cells generated an amplicon of 140 bp (Figure 1C). In contrast, these amplicons were not found in SMMC-7721 cells and SMMC-7721/pcDNA3.1+ cells, demonstrating that the gene of anti-hVEGF hairpin ribozyme transfected into SMMC-7721 cells could be integrated with SMMC-7721 cell genome and transcripted into corresponding mRNA efficiently.
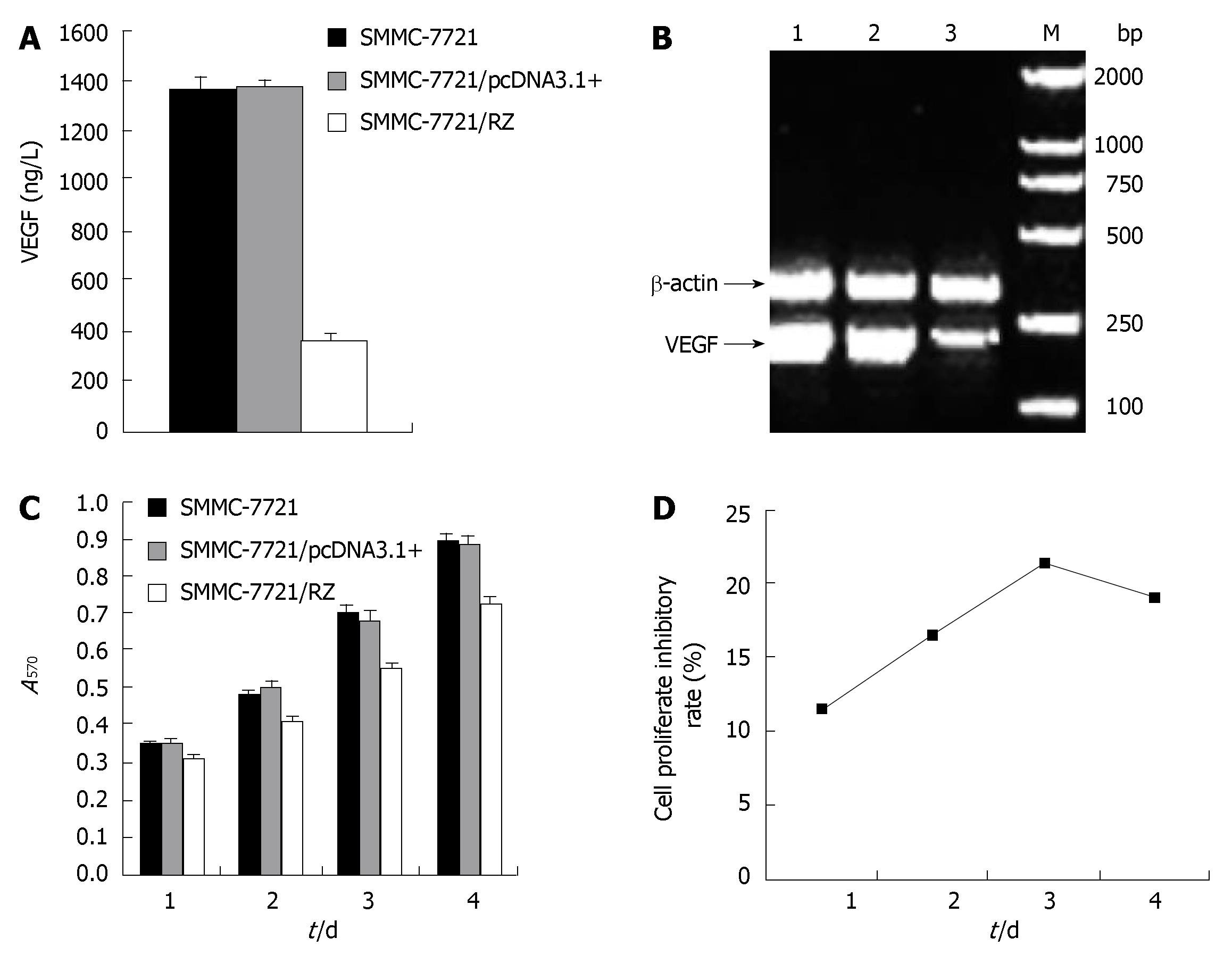
As the ultimate goal we sought to achieve was the reduction of VEGF secretion into extracellular compartment by SMMC-7721 cells, we investigated if transfection of ribozyme has a direct effect on VEGF secretion by SMMC-7721 cells. As demonstrated in Figure 2A, VEGF protein expression level detected by ELISA in transgenic SMMC-7721/RZ group was 363.64 ± 19.68 ng/L, which was significantly lower than that in SMMC-7721 cell group (1358.69 ± 49.81 ng/L) and SMMC-7721/pcDNA3.1+ group (1369.57 ± 32.61 ng/L) (P < 0.01). There was no significant difference between SMMC-7721 cell group and SMMC-7721/pcDNA3.1+ group (P > 0.05). These results indicate that ribozyme inhibited VEGF expression (more than 73%) in HCC cells in vitro.
To determine whether the inhibitory effect of ribozyme on VEGF expression is at the transcriptional level, VEGF mRNA expression in tumor cells was detected by RT-PCR. The products were run on 1% agarose gels and visualized with ethidium bromide staining. SMMC-7721/RZ cells and two control group cells generated an amplicon of 195 bp. By normalizing to β-actin, the level of VEGF mRNA expression in SMMC-7721/RZ cells was remarkably lower than that in two control groups (Figure 2B), suggesting that ribozyme could reduce VEGF expression by inhibiting its transcription.
As shown in Figure 2C-D, after cells were cultured for 24, 48, 72 and 96 h, the proliferation rate for SMMC-7721/RZ cells was significantly lower than that for SMMC-7721 cells and SMMC-7721/pcDNA3.1+ cells (P < 0.01). On the third day, ribozyme-mediated growth inhibition rate reached 78.6%. There was no significant difference in proliferation of SMMC-7721 cells and SMMC-7721/pcDNA3.1+ cells (P > 0.05). The results reveal that ribozyme inhibited proliferation of HCC cells in vitro.
To determine the impact of anti-hVEGF hairpin ribozyme on tumorigenicity in vivo, control and ribozyme-transduced cells were transplanted subcutaneously into BLAB/c nude mice. Histology and immunohistochemistry displayed different levels of VEGF expression and angiogenesis on sections of xenografted and control tumor tissues. As expected, tumors appeared on d 14 (± 6.3) and d 7 (± 2.6) in SMMC-7721/RZ group and SMMC-7721/pcDNA3.1+ group, respectively (P < 0.01). The ribozyme transgenic cells resulted in tumor formation with lower levels of VEGF expression and angiogenesis and delayed tumor formation on 17.2 ± 13.7 d compared with 8.6 ± 3.3 d in control cells (P < 0.01). Formation of large, red (vascular) tumors was observed in SMMC-7721/pcDNA3.1+ cells of all mice. However, formation of small white (avascular) tumors was found after inoculation with SMMC-7721/RZ cells. The tumor volume and weight at the time of sacrifice in SMMC-7721/RZ group (0.19 ± 0.0085 cm3, 0.26 ± 0.076 g) were significantly lower than those in SMMC-7721/pcDNA3.1+ group (0.59 ± 0.019 cm3, 0.74 ± 0.050 g, P < 0.01). The inhibitory effects of anti-hVEGF hairpin ribozyme on HCC xenograft growth in nude mice were comparable and significant (P < 0.01).
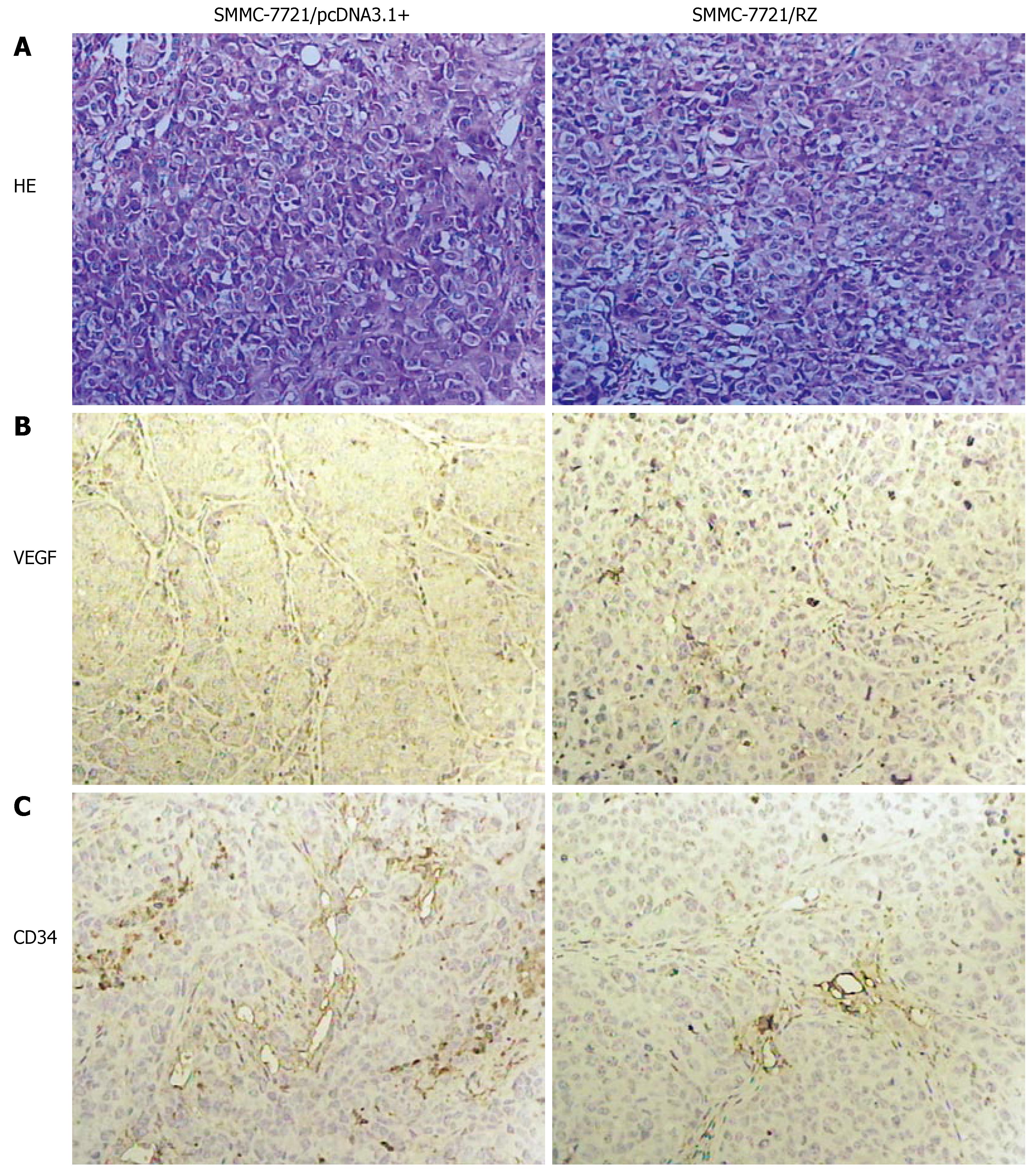
As shown in Figure 3A, different shape and size of tumor cells were found in SMMC-7721/pcDNA3.1+ group. Their nuclei were large with a disproportional ratio of karyoplasm. Nuclear chromatin was condensed. Several basophilia chromatospherites were seen. Normal karyokinesis (8.26 ± 0.55/visual field) and pathologic karyokinesis (8.52 ± 0.17/visual field) were frequently observed. In contrast, the shape and size of tumor cells in SMMC-7721/RZ group were coincident. Nuclear chromatin was puff. Normal karyokinesis (3.18 ± 0.23/visual field) and pathologic karyokinesis (3.3 ± 0.37/visual field) were significantly less than those in SMMC-7721/pcDNA3.1+ group (P < 0.01).
Immunohistochemical staining showed VEGF as light brown particles. VEGF chromatosis was distinctly less or/and weaker in SMMC-7721/RZ group than in SMMC-7721/pcDNA3.1+ group (Figure 3B).
The control mice displayed well vascularized tumors. Tumor blood vessels and their wall were abundant in the control group. In SMMC-7721/RZ group, however, tumor blood vessels were seldom seen with vessel wall frame collapsed. As shown in Figure 3C, a significant difference in the relative intensity of immunostaining for anti-CD34 antibody (an endothelial marker) was observed between the SMMC-7721/RZ group and the control group, indicating that vascularization was decreased in SMMC-7721/RZ group after transfection of anti-hVEGF hairpin ribozyme. MVD in the sections was significantly lower in SMMC-7721/RZ group (3.82 ± 0.88) than in the control group (7.02 ± 0.14, P < 0.01).
It was reported that ribozyme is used to inhibit cellular targets[23-25]. Among them is the ribozyme-based therapeutics for cancer which might be devised to inhibit tumor growth or to prevent its metastasis. Compared to other gene technologies, the superiorities of ribozyme are as follows: (1) ribozyme has dual effects of cleavage and blockade, (2) it cannot be easily hydrolyzed by nucleic acid enzymes because it can bind to target RNA to form a stable helix structure, (3) ribozyme molecule can destruct multiple target RNAs and be used repeatedly[26]. Biochemical characterization has shown that hairpin ribozyme is one of the most efficient ribozymes.
In order to devise effective anti-hVEGF hairpin ribozymes, we performed a sequence analysis of hVEGF mRNA and studied its secondary structure. We took into account the ideal site of hairpin ribozyme cleavage and its substrates in exon 3 of a GUC since this site is more easy for hairpin ribozyme to approach and less affected by the secondary structure. A fragment of the ribozyme gene and its sequence were verified by digestion of pcDNA3.1+/RZ with BamHI and EcoRI and sequencing. Our results show that the recombinant eukaryotic expression vector pcDNA3.1+/RZ could be successfully constructed. Stable integration and transcription of the ribozyme gene in SMMC-7721 cells were confirmed by PCR and RT-PCR. When introduced into hepatocarcinoma cell line SMMC-7721, anti-hVEGF hairpin ribozyme suppressed not only VEGF mRNA transcription but also VEGF protein expression. VEGF protein expression was inhibited by approximately 73% in SMMC-7721 cells transfected by ribozyme. These results reveal that the constructed ribozyme can selectively inhibit VEGF gene expression in human hepatocarcinoma cells. Ciafre et al[27] have developed an anti-VEGF hammerhead ribozyme targeting the 5' part of human VEGF mRNA. Transfection of U87 human glioblastoma cells with plasmid vectors encoding for this ribozyme resulted in a strong (-56%) reduction of VEGF secretion in the extracellular medium. We believe that our ribozyme has a better biological activity in selectively inhibiting VEGF expression. The successful use of ribozymes as therapeutic agent depends upon many factors, e.g., category of ribozymes (hairpin and hammerhead ribozymes), relative amount of active ribozymes in cells, their co-localization with target RNAs, structural features of transcripts influencing accessibility to specific target sites, catalytic efficiency, interaction of target RNAs with proteins, and intracellular stability of targeted RNAs and ribozymes.
Little is known about the role of VEGF in HCC cell proliferation, differentiation and tumorigenesis. One of the more intriguing aspects of our results is that after cells were cultured for 24, 48, 72 and 96 h, the proliferation rate for SMMC-7721/RZ cells was significantly lower than that for SMMC-7721 cells and blank vector cells, and the inhibitory rate of ribozyme for the growth of SMMC-7721 cells was 21.4% on the third day. Apoptosis peak was detected by flow cytometry in SMMC-7721/RZ cells, the rate of apoptosis was 11.0% (data not shown), suggesting that the expression of anti-hVEGF hairpin ribozyme can significantly reduce VEGF secretion by transfected cells and inhibit cell growth and apoptosis in hepatocarcinoma cells. In other words, VEGF signaling can prolong the survival of HCC cells that may be independent of angiogenesis. Since Masood et al[28] reported that the expression of Flt-1 and KDR, the receptors of VEGF, is high in AIDS-Kaposi sarcoma cells and primary tumor tissues. Similar results have been observed in several tumors, by researchers in succession[29-31]. To inhibit VEGF expression or to block VEGF binding to its receptor results in growth inhibition of tumor cell lines, such as melanoma and ovarian carcinoma cell lines expressing VEGF receptors. In sharp contrast, cell lines not expressing VEGFRs show no response[28]. VEGF addition causes phosphorylation of mitogen-activated protein kinase as well as VEGF receptor, and induces proliferation and migration of lung cancer cells[32]. Our results are consistent with those previously observed. These findings suggest that tumor cells secrete VEGF which not only induces angiogenesis but also acts as an autocrine growth factor for carcinoma cells defined by the expression of VEGF receptors[33]. Our data suggest that HCC cells express specific VEGF receptors which respond to autocrine VEGF, thus activating signaling pathways that impede apoptosis and promote cell proliferation.
Because results on cell lines often represent a distorted and incomplete picture of the in situ physiopathology of cancer where the tumor microenvironment and neovascularization play a critical role in tumor growth and progression, we expanded our study to matched primary tumors using xenograft models. Oncogenicity is a critical index to judge malignant proliferation. The transfected SMMC-7721 cells were transplanted into nude mice. VEGF expression and microvascularization were distinctly decreased in SMMC-7721/RZ cell group when compared with SMMC-7721/pcDNA3.1+ cell group. Furthermore, tumor formation was delayed and its growth was significantly slowed down by ribozyme. Furthermore, reduced VEGF production induced by ribozyme resulted in higher cell differentiation, less cyto-heteromorphism and proliferation in the ribozyme transfected group, as demonstrated by microscopy, suggesting that VEGF is functionally related to cell differentiation directly or indirectly by regulating expression of certain genes. Our data reveal that VEGF plays a basic role in the induction of tumorigenicity, and underlines the importance of inhibiting its production. Moreover, Xu et al[34] found that down-regulation of VEGF gene by introducing anti-VEGF hairpin ribozyme gene into leukemia cell line K562 can alter the gene expression profiles. Among the 4096 gene clones on the microarray, 191 were detected to have the marked changes with 104 down-regulated and 87 up-regulated, showing that they are functionally related to cell cycle progression, gene replication, metabolism, cell apoptosis, cell signal transduction, and oncogenes[34]. All these findings indicate that VEGF is a key molecule for tumor progression, although it is unclear how VEGF expression regulates various genes.
In human hepatocarcinoma tumor xenograft expe-riment, although there was a significant difference in P value between the control and ribozyme groups, tumor formation was observed in the ribozyme group. To further decrease tumor formation, in combined with anti-VEGF ribozyme, it is necessary to explore ribozymes targeting other genes, which contribute to hepatocarcinoma tumorigenesis such as N- ras, C-myc and IGF-II.
In conclusion, VEGF promotes tumorigenesis and angiogenesis, and anti-hVEGF hairpin ribozyme can efficiently inhibit the VEGF expression and growth of hepatocarcinoma in vitro and in vivo. In VEGFRs-expressing tumors such as HCC, VEGF inhibition may be attributed to inhibition of tumor angiogenesis and direct effects on tumor cell proliferation/survival. Anti-hVEGF hairpin ribozyme may be a candidate for HCC gene therapy. However, its clinical application needs further study.
Angiogenesis, known as the development and proliferation of new blood vessels, is vital for the growth of tumors. Vascular endothelial growth factor (VEGF) is an essential cytokine in the regulation of angiogenesis. Little is known about the role of VEGF in hepatocellular carcinoma (HCC) proliferation, differentiation and tumorigenesis. The present study was to study the effectiveness and mechanisms of anti-human VEGF (hVEGF) hairpin ribozyme on VEGF expression, oncogenicity and tumor growth in hepatocarcinoma cell line and xenografted model.
HCC is a highly vascular tumor depending on neovascularization and insensitive conventional chemotherapeutics. It is important to explore antiangiogenesis methods for HCC. Hairpin ribozyme has been used in gene therapy.
To our knowledge, this is the first report that anti-hVEGF hairpin ribozyme can effectively inhibit VEGF expression and growth of hepatocarcinoma in vitro and in vivo. VEGF inhibition can inhibit tumor angiogenesis and tumor growth that may be dependent and independent of angiogenesis in HCC. Our results also suggest that VEGF is functionally related to cell proliferation, differentiation and tumorigenesis.
Anti-hVEGF hairpin ribozyme can be used as a therapeutic agent for hepa-tocarcinoma.
Ribozymes: a group of catalytically active nucleic acids capable of site-specific cleavage of target mRNAs, thus decreasing mRNA expression and inhibiting the function of target gene.
In this paper, the role of anti-VEGF hairpin ribozyme in antitumor and antigangiogenic activities is described. The experiments were well designed. The data support that VEGF plays its role through angiogenesis and other pathways directly leading to carcinoma.
S- Editor Zhu LH L- Editor Wang XL E- Editor Ma WH
| 1. | Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6437] [Cited by in RCA: 6484] [Article Influence: 259.4] [Reference Citation Analysis (0)] |
| 2. | Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3:65-71. [PubMed] |
| 3. | Ribatti D. The crucial role of vascular permeability factor/vascular endothelial growth factor in angiogenesis: a historical review. Br J Haematol. 2005;128:303-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 130] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc Health Risk Manag. 2006;2:213-219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 804] [Cited by in RCA: 1021] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 5. | Yao DF, Wu XH, Zhu Y, Shi GS, Dong ZZ, Yao DB, Wu W, Qiu LW, Meng XY. Quantitative analysis of vascular endothelial growth factor, microvascular density and their clinicopathologic features in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2005;4:220-226. [PubMed] |
| 6. | Iavarone M, Lampertico P, Iannuzzi F, Manenti E, Donato MF, Arosio E, Bertolini F, Primignani M, Sangiovanni A, Colombo M. Increased expression of vascular endothelial growth factor in small hepatocellular carcinoma. J Viral Hepat. 2007;14:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Horrée N, van Diest PJ, van der Groep P, Sie-Go DM, Heintz AP. Hypoxia and angiogenesis in endometrioid endometrial carcinogenesis. Cell Oncol. 2007;29:219-227. [PubMed] |
| 8. | Möbius C, Demuth C, Aigner T, Wiedmann M, Wittekind C, Mössner J, Hauss J, Witzigmann H. Evaluation of VEGF A expression and microvascular density as prognostic factors in extrahepatic cholangiocarcinoma. Eur J Surg Oncol. 2007;33:1025-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | El-Gohary YM, Silverman JF, Olson PR, Liu YL, Cohen JK, Miller R, Saad RS. Endoglin (CD105) and vascular endothelial growth factor as prognostic markers in prostatic adenocarcinoma. Am J Clin Pathol. 2007;127:572-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 10. | Hlatky L, Tsionou C, Hahnfeldt P, Coleman CN. Mammary fibroblasts may influence breast tumor angiogenesis via hypoxia-induced vascular endothelial growth factor up-regulation and protein expression. Cancer Res. 1994;54:6083-6086. [PubMed] |
| 11. | Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3206] [Cited by in RCA: 3224] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 12. | Gratzinger D, Zhao S, Marinelli RJ, Kapp AV, Tibshirani RJ, Hammer AS, Hamilton-Dutoit S, Natkunam Y. Microvessel density and expression of vascular endothelial growth factor and its receptors in diffuse large B-cell lymphoma subtypes. Am J Pathol. 2007;170:1362-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Kopfstein L, Veikkola T, Djonov VG, Baeriswyl V, Schomber T, Strittmatter K, Stacker SA, Achen MG, Alitalo K, Christofori G. Distinct roles of vascular endothelial growth factor-D in lymphangiogenesis and metastasis. Am J Pathol. 2007;170:1348-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 14. | Nikiteas NI, Tzanakis N, Theodoropoulos G, Atsaves V, Christoni Z, Karakitsos P, Lazaris AC, Papachristodoulou A, Klonaris C, Gazouli M. Vascular endothelial growth factor and endoglin (CD-105) in gastric cancer. Gastric Cancer. 2007;10:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Djordjevic G, Mozetic V, Mozetic DV, Licul V, Ilijas KM, Mustac E, Oguic R, Fuckar Z, Jonjic N. Prognostic significance of vascular endothelial growth factor expression in clear cell renal cell carcinoma. Pathol Res Pract. 2007;203:99-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Byrne AM, Bouchier-Hayes DJ, Harmey JH. Angiogenic and cell survival functions of vascular endothelial growth factor (VEGF). J Cell Mol Med. 2005;9:777-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 523] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 17. | Orito E, Mizokami M. Hepatitis B virus genotypes and hepatocellular carcinoma in Japan. Intervirology. 2003;46:408-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Tabor E. Tumor suppressor genes, growth factor genes, and oncogenes in hepatitis B virus-associated hepatocellular carcinoma. J Med Virol. 1994;42:357-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 76] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Jiang YF, Yang ZH, Hu JQ. Recurrence or metastasis of HCC: predictors, early detection and experimental antiangiogenic therapy. World J Gastroenterol. 2000;6:61-65. [PubMed] |
| 20. | Toi M, Matsumoto T, Bando H. Vascular endothelial growth factor: its prognostic, predictive, and therapeutic implications. Lancet Oncol. 2001;2:667-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 179] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 21. | Puerta-Fernández E, Romero-López C, Barroso-delJesus A, Berzal-Herranz A. Ribozymes: recent advances in the development of RNA tools. FEMS Microbiol Rev. 2003;27:75-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Yan RL, Qian XH, Xin XY, Jin M, Hui HX, Wang DT, Wang J. Experimental study of anti-VEGF hairpin ribozyme gene inhibiting expression of VEGF and proliferation of ovarian cancer cells. Ai Zheng. 2002;21:39-44. [PubMed] |
| 23. | Vashishta A, Ohri SS, Proctor M, Fusek M, Vetvicka V. Ribozyme-targeting procathepsin D and its effect on invasion and growth of breast cancer cells: an implication in breast cancer therapy. Int J Oncol. 2007;30:1223-1230. [PubMed] |
| 24. | Li QX, Tan P, Ke N, Wong-Staal F. Ribozyme technology for cancer gene target identification and validation. Adv Cancer Res. 2007;96:103-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Jung HS, Lee SW. Ribozyme-mediated selective killing of cancer cells expressing carcinoembryonic antigen RNA by targeted trans-splicing. Biochem Biophys Res Commun. 2006;349:556-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Rossi JJ. Controlled, targeted, intracellular expression of ribozymes: progress and problems. Trends Biotechnol. 1995;13:301-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Ciafrè SA, Niola F, Wannenes F, Farace MG. An anti-VEGF ribozyme embedded within the adenoviral VAI sequence inhibits glioblastoma cell angiogenic potential in vitro. J Vasc Res. 2004;41:220-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 28. | Masood R, Cai J, Zheng T, Smith DL, Naidu Y, Gill PS. Vascular endothelial growth factor/vascular permeability factor is an autocrine growth factor for AIDS-Kaposi sarcoma. Proc Natl Acad Sci USA. 1997;94:979-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 217] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 29. | Masood R, Cai J, Zheng T, Smith DL, Hinton DR, Gill PS. Vascular endothelial growth factor (VEGF) is an autocrine growth factor for VEGF receptor-positive human tumors. Blood. 2001;98:1904-1913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 250] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 30. | Ng IO, Poon RT, Lee JM, Fan ST, Ng M, Tso WK. Microvessel density, vascular endothelial growth factor and its receptors Flt-1 and Flk-1/KDR in hepatocellular carcinoma. Am J Clin Pathol. 2001;116:838-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 127] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Yoshiji H, Kuriyama S, Hicklin DJ, Huber J, Yoshii J, Ikenaka Y, Noguchi R, Nakatani T, Tsujinoue H, Fukui H. The vascular endothelial growth factor receptor KDR/Flk-1 is a major regulator of malignant ascites formation in the mouse hepatocellular carcinoma model. Hepatology. 2001;33:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Tanno S, Ohsaki Y, Nakanishi K, Toyoshima E, Kikuchi K. Human small cell lung cancer cells express functional VEGF receptors, VEGFR-2 and VEGFR-3. Lung Cancer. 2004;46:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 97] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 33. | Mercurio AM, Bachelder RE, Bates RC, Chung J. Autocrine signaling in carcinoma: VEGF and the alpha6beta4 integrin. Semin Cancer Biol. 2004;14:115-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Xu WL, Shen HL, Wu ZY, Tang HR, Wang FC. The down regulation of VEGF gene expression may link to change the expression profile of genes in leukemia cell line K562. Zhonghua YiXue YiChuanXue ZaZhi. 2006;23:37-42. [PubMed] |