Published online Dec 14, 2007. doi: 10.3748/wjg.v13.i46.6236
Revised: September 20, 2007
Accepted: October 29, 2007
Published online: December 14, 2007
AIM: To confirm the predictive factors for interferon (IFN)-α and ribavirin combination therapy for chronic hepatitis patients with hepatitis C virus (HCV) genotype 1b.
METHODS: HCV RNA from 50 patients infected with HCV genotype 1b was studied by cloning and sequencing of interferon sensitivity determining region (ISDR), PKR-eIF2α phosphorylation homology domain (PePHD). Patients were treated with IFN-α and ribavirin for 6 mo and grouped by effectiveness of the therapy. A variety of factors were analyzed.
RESULTS: Our data showed that age, HCV RNA titer, and ISDR type could be used as the predictive factors for combined IFN-α and ribavirin efficacy. Characteristically, mutations in PePHD appeared only when the combination therapy was effective. Other factors, such as sex and alanine aminotransferase (ALT) level, were not related to its efficacy. Adjusting for age and HCV RNA titer indicated that the ISDR type was the most potent predictive factor.
CONCLUSION: HCV RNA ISDR type is an important factor for predicting efficacy of IFN-α and ribavirin combination therapy in Korean patients.
- Citation: Yoon J, Lee JI, Baik SK, Lee KH, Sohn JH, Lee HW, Namkung J, Chang SJ, Choi JW, Kim HW, Yeh BI. Predictive factors for interferon and ribavirin combination therapy in patients with chronic hepatitis C. World J Gastroenterol 2007; 13(46): 6236-6242
- URL: https://www.wjgnet.com/1007-9327/full/v13/i46/6236.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i46.6236
Hepatitis C virus (HCV) is an enveloped RNA virus with a positive single-stranded RNA genome about 9600 nucleotides in length. This RNA encodes a single polypeptide approximately 3000 amino acids in length. The polypeptide is post-translationally cleaved into structural and non-structural proteins[1-3]. HCV infection is a major cause for chronic liver disease worldwide[4]. Eighty percent or more of acutely infected patients develop chronic hepatitis, which progresses to liver cirrhosis in about 20% of cases and hepatocellular carcinoma in 5%. These complications arise even if the disease remains asymptomatic[5].
Alpha-interferon (IFN-α) treatment effectively reduces viral load, but complete eradication of the virus is achieved in less than 20% of patients treated with IFN-α alone and in 40%-47% of patients treated with combined IFN-α and ribavirin[6-8]. The treatment outcome largely depends on the sensitivity of HCV genotype and IFN-α[9,10]. About 10 years before, IFN-α and ribavirin combination therapy has begun to be used instead of IFN monotherapy. Ribavirin is an oral nucleoside analogue acting on a broad spectrum of DNA and RNA viruses. It has been proposed to have both direct antiviral and immunomodulatory effects[11], but the detailed mechanism remains unclear. Ribavirin used as monotherapy is known to have little or no activity against HCV.
The HCV genotype appears to be a major determinant for IFN efficacy, because patients infected with HCV genotypes 2 and 3 respond better to IFN monotherapy than patients with genotype 1[9]. The IFN-based therapy effectiveness is not satisfactory, especially in patients with HCV genotype 1b[12-14], the most common genotype in Korea[15,16], Japan[17], southern and eastern Europe[18,19]. IFN is also inconvenient to use, costly, and has a variety of possible complications. Thus, many studies have been carried out to determine the predictive factors for the efficacy IFN therapy[12-19].
Response to interferon monotherapy is associated with several host and viral factors. HCV genotype 1b, low viral load, and rapid HCV RNA clearance from the serum have been identified as favorable predictors for a sustained response to IFN therapy[20-22]. Since Enomoto et al[23] reported that genetic variability in a 40 amino acid stretch (amino acids 2209-2248) and mutations in the NS5A region of HCV and in designated interferon sensitivity determining region (ISDR), has become a predictive factor for IFN therapy. Studies from Japan[23], Sweden[24] and Spain[25] have shown that ISDR is an effective predictor. However, studies from Western countries displayed that ISDR is not a good predictor[26-28]. Studies on the relationship between IFN-α and ribavirin combination therapy and ISDR have controversial results[29-32].
Taylor et al[31] reported that a HCV envelope protein (E2) contains a sequence similar to the phosphorylation site on eIF2-α for the interferon-inducible cellular protein kinase PKR. The PKR-eIF2-α phosphorylation homology domain (PePHD) on E2 may serve as a pseudosubstrate for PKR and inhibit its function, reducing the antiviral effect of interferon. Thus, the PePHD region might also be involved in IFN resistance of chronic hepatitis C to IFN therapy. However, the role of this region is also controversial so far[31].
In Korea, the HCV prevalence is about 1%-2%, but studies to analyze the predictive factors for combined IFN-α and ribavirin therapy have not been performed. To identify these factors, we investigated the relationship between combined IFN-α and ribavirin efficacy and a variety of factors such as ISDR sequence, PePHD sequence, age, ALT level, and HCV RNA titer in Korean patients with HCV genotype 1b.
Serum was collected from HCV genotype 1b-infected patients admitted to Wonju Christian Hospital. Only HCV genotype 1b was used in this study because it is the most common HCV genotype in the Republic of Korea. Sera were screened by a third generation ELISA method with an anti-HCV antibody. The patients were treated with IFN-α and ribavirin for 6 mo. Three million units of IFN-α was injected every two days, and 9 mg of ribavirin was orally administrated during the same period. The patients who did not receive the treatment were excluded. After the 6-mo combination therapy, the patients were classified into complete response group and no-response group. In the complete response group, HCV RNA titer was less than 50 IU/mL and ALT levels were within the normal range. In the no-response group, HCV RNA titer was over 50 IU/mL even if the ALT levels were normal.
HCV RNA was extracted from sera as previously desc-ribed[33]. After ethanol precipitation, each RNA pellet was dissolved in 10 μL of diethylpyrocarbonate (DEPC)-treated distilled water for cDNA preparation. cDNA synthesis was performed as previously described[34] with certain modifications. For the synthesis of cDNA of HCV, an aliquot of RNA (10 μL) isolated from the sera of patients was mixed with 1 μL of random hexamer (1 μmol/L), 2 μL of reaction buffer (250 mmol/L Tris-HCl pH 8.3, 250 mmol/L potassium chloride, 50 mmol/L magnesium chloride, 50 mmol/L dithiothreitol and 2.5 mmol/L spermidine) and 5.5 μL of DEPC-treated water was added. After the contents were heat-treated for 5 min at 65°C, 20 units (0.5 μL) of RNase inhibitor and 10 units (1 μL) of AMV reverse transcriptase were added. The mixture was incubated at 37°C for 30 min, followed by at 99°C for 1 min to inactivate the enzyme. PCR was performed as described previously[16]. The ISDR and PePHD primer sequences are listed in Table 1. PCR products were subjected to agarose gel electrophoresis in Tris-acetate-EDTA buffer and visualized with ethidium bromide staining under an ultraviolet transilluminator.
| Region | Primer direction | Sequence (5’ to 3’) | Nucleotide No |
| ISDR | Outer sense | TGGATGGAGTGCGGTTGCACAGGTA | 6703-6727 |
| Outer antisense | TGTAAAACGACGGCCAG | 7296-7320 | |
| Inner sense | TCTTTCTCCGTGGAGGTGGTATTGC | 6722-6741 | |
| Inner antisense | CAGGAAACAGCTATGACC | 7275-7294 | |
| PePHD | Outer sense | TGACTACCCATACAGGCTCT | 2180-2199 |
| Outer antisense | AAGGAAGGAGAGATTGCCAT | 2725-2744 | |
| Inner sense | AAGGTTAGGATGTATGTGGG | 2238-2257 | |
| Inner antisense | ATTGAGGACCACCGAGTTCT | 2689-2708 |
RT-PCR amplified products, including the ISDR and/or PePHD regions, were purified from agarose gel and glass milk (Gene Clean II kit, Bio 101, USA), and then subcloned by inserting the cDNA into a pGEM-T TA-cloning vector (Promega). The clones from each of the individual patient's plates were randomly selected and plasmid prepared from each clone was used as a template for DNA sequencing which was performed as previously described[35].
In order to determine the HCV RNA titer, a quantitative and competitive polymerase chain reaction (QCPCR) assay was carried out as previously described[36]. As a first step, cDNA encoding the 5'-untranslated region of HCV was subcloned into a pGEM vector (pGEM5'UTR). Using PCR, the internal control plasmid, pGEM5'UTRDel, was constructed by deletion of nucleotides between the 87 and 165 nucleotides in the 5'-UTR of the HCV genome. The internal control RNA was synthesized in vitro by T7 RNA polymerase from a linearized template derived from the pGEM5'UTRDel plasmid. The amount of RNA synthesized in vitro was determined by measurement of the absorbance at 260 nm. A known copy number of the RNA was included as an internal control in order to quantify the viral RNA. The data were analyzed by Quantity One® 1-D analysis software (Bio-Rad).
Comparisons between groups were made by the Student's t-test. The P values were determined between the two groups with regard to age, ALT, amino acid mutations in PePHD, and HCV RNA titer. P < 0.05 was considered statistically significant. Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated for ISDR to test its predictive value for combination therapy by logistic regression analysis.
We collected serum from 50 HCV genotype 1b-infected patients. Among them, 21 patients completely responded to the 6-mo combination therapy, while 29 patients showed no response. Group A was designated as the complete response group and group B as the no-response group. The characteristics of each patient prior to combination therapy are shown in Tables 2 and 3.
| Age(yr) | Sex | ALT | Transfusionhistory1 | HCV RNAtiter2 | ISDRType3 | No. of AminoAcid Mutationsof PePHD | |
| A-1 | 50 | M | 178 | × | 3.92 | 1 | 0 |
| A-2 | 49 | F | 135 | ○ | 3.27 | 1 | 1 |
| A-3 | 40 | M | 106 | × | 4.16 | 1 | 1 |
| A-4 | 45 | M | 218 | ○ | 5.48 | 1 | 0 |
| A-5 | 51 | F | 92 | × | 4.04 | 1 | 0 |
| A-6 | 41 | M | 143 | × | 3.75 | 2 | 0 |
| A-7 | 56 | F | 167 | × | 4.24 | 2 | 0 |
| A-8 | 58 | M | 86 | × | 5.26 | 2 | 0 |
| A-9 | 49 | M | 99 | ○ | 5.12 | 2 | 1 |
| A-10 | 40 | F | 129 | ○ | 4.41 | 2 | 0 |
| A-11 | 54 | F | 201 | × | 3.73 | 2 | 1 |
| A-12 | 53 | M | 179 | × | 4.21 | 2 | 1 |
| A-13 | 52 | M | 234 | × | 4.25 | 2 | 0 |
| A-14 | 47 | M | 311 | × | 5.11 | 2 | 0 |
| A-15 | 49 | M | 86 | × | 6.12 | 3 | 1 |
| A-16 | 53 | F | 220 | × | 5.91 | 3 | 0 |
| A-17 | 47 | F | 194 | × | 4.82 | 3 | 0 |
| A-18 | 49 | M | 246 | × | 5.44 | 3 | 1 |
| A-19 | 51 | M | 441 | ○ | 5.22 | 3 | 2 |
| A-20 | 47 | F | 305 | ○ | 4.85 | 3 | 0 |
| A-21 | 55 | M | 119 | × | 3.73 | 3 | 0 |
| Age(yr) | Sex | ALT | Transfusionhistory1 | HCV RNAtiter2 | ISDRtype3 | No. of AminoAcid Mutationsof PePHD | |
| B-1 | 53 | M | 185 | × | 5.25 | 1 | 0 |
| B-2 | 48 | M | 320 | × | 6.11 | 1 | 0 |
| B-3 | 44 | F | 125 | ○ | 4.88 | 1 | 0 |
| B-4 | 49 | F | 175 | × | 5.72 | 1 | 0 |
| B-5 | 54 | M | 151 | ○ | 6.21 | 1 | 0 |
| B-6 | 58 | F | 190 | × | 5.14 | 1 | 0 |
| B-7 | 62 | F | 212 | × | 6.35 | 1 | 0 |
| B-8 | 64 | M | 252 | ○ | 7.11 | 1 | 0 |
| B-9 | 45 | M | 145 | × | 6.82 | 1 | 0 |
| B-10 | 49 | M | 138 | × | 4.95 | 1 | 0 |
| B-11 | 42 | F | 120 | ○ | 5.54 | 1 | 0 |
| B-12 | 55 | M | 95 | × | 6.25 | 1 | 0 |
| B-13 | 53 | M | 142 | × | 6.73 | 1 | 0 |
| B-14 | 55 | F | 185 | × | 6.76 | 1 | 0 |
| B-15 | 57 | M | 258 | × | 4.85 | 1 | 0 |
| B-16 | 53 | F | 175 | ○ | 6.33 | 2 | 0 |
| B-17 | 49 | M | 241 | × | 5.81 | 2 | 0 |
| B-18 | 44 | M | 183 | × | 6.32 | 2 | 0 |
| B-19 | 59 | F | 167 | × | 5.89 | 2 | 0 |
| B-20 | 54 | M | 171 | ○ | 6.14 | 2 | 0 |
| B-21 | 52 | F | 217 | × | 6.23 | 2 | 0 |
| B-22 | 55 | M | 222 | × | 5.88 | 2 | 0 |
| B-23 | 63 | F | 235 | × | 6.32 | 2 | 0 |
| B-24 | 47 | F | 161 | × | 6.47 | 2 | 0 |
| B-25 | 51 | M | 96 | ○ | 5.31 | 2 | 0 |
| B-26 | 54 | F | 80 | × | 5.85 | 2 | 0 |
| B-27 | 55 | F | 192 | ○ | 6.42 | 3 | 0 |
| B-28 | 58 | M | 234 | × | 5.93 | 3 | 0 |
| B-29 | 48 | M | 341 | × | 4.98 | 3 | 0 |
As shown in Tables 2 and 3, the HCV RNA titer had a wide distribution. In the complete response group the HCV RNA titer was between 103.27-106.12 copies per mL and 104.85-107.11 copies per mL, respectively, in the no-response group. The average RNA titer of the response and no-response groups was 4.62 ± 0.80 and 5.59 ± 0.61, respectively. These values were statistically significant (P < 0.05, Table 4).
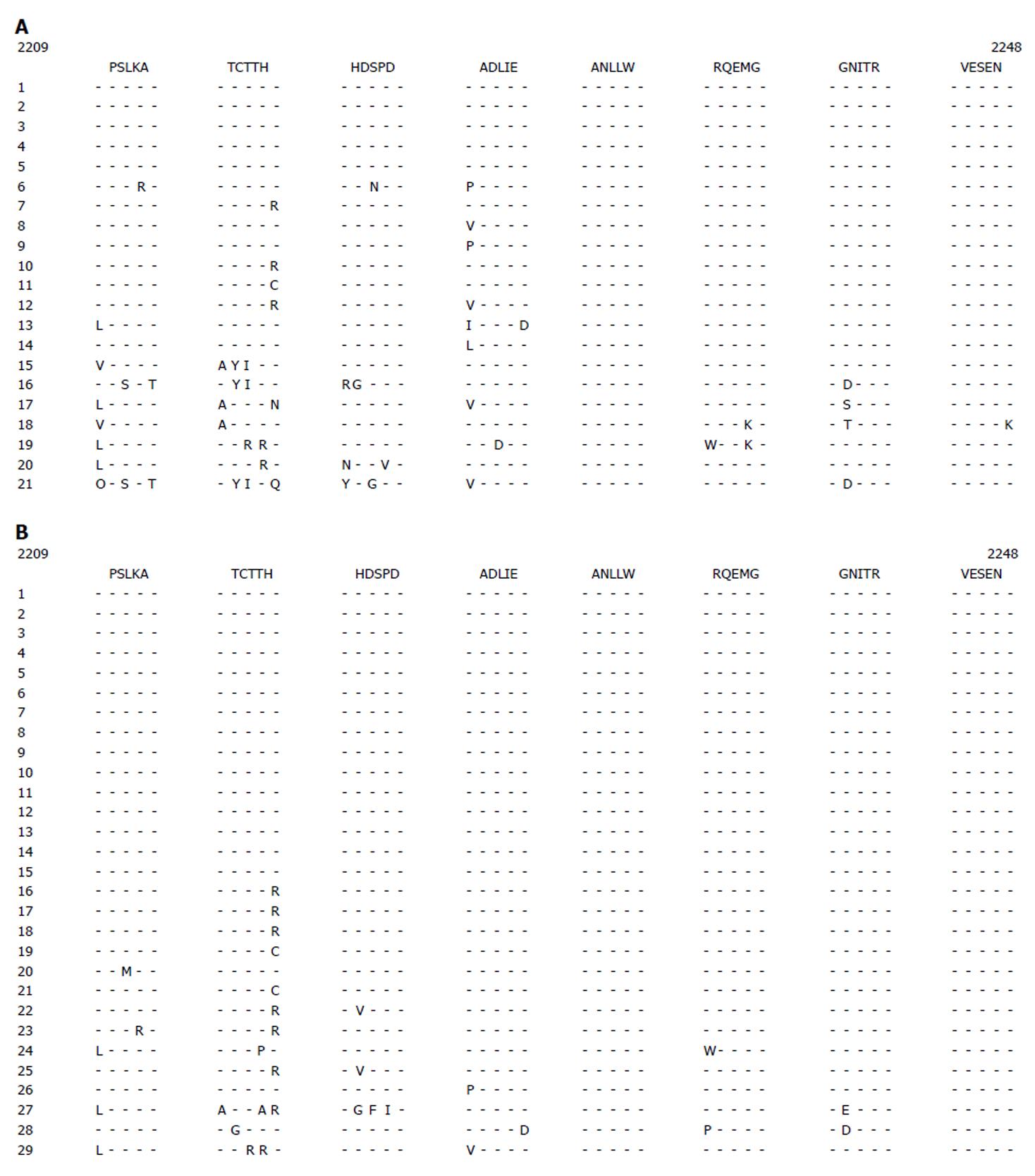
The ISDR and PePHD amino acid sequences and the HCV genotype 1b prototype sequence (HCV-J) are shown in Figure 1. The complete response group had 1-10 amino acid substitutions while the no-response group had 1-8 amino acid substitutions in the ISDR (Figure 1A and B). The PePHD region had 1-2 amino acid substitutions in several cases of complete response group and no amino acid substitutions in the no-response group (Figure 2).
As shown in Table 4, patients were younger in the complete response group than in the no-response group. The HCV RNA titer was also significantly different. No differences were found in gender or ALT levels. We classified ISDR sequences into three groups based on the number of amino acid mutations, as previously described[23]. These three groups were analyzed by an odds ratio to define the responsiveness to combination therapy. The ISDR group responses to combination therapy are shown in Table 5. Intermediate (one to three amino acid changes) and mutant (four or more amino acid changes) ISDRs showed an increased responsiveness to the combination therapy. The odds ratio was 2.46 and 7.00, respectively, assuming the wild type had 1.00. The age, PePHD mutations, and HCV RNA titer were significantly different between the two groups (Table 4). Considering these factors, ISDR type might be a better predictive factor for combination therapy responsiveness. After adjusting for age and HCV RNA titer, the odds ratio for intermediate and mutant ISDRs was 3.57 and 9.67, respectively. According to these results, patients with mutant ISDR strains would likely respond better to combination therapy than those with wild type ISDR strains.
Identifying host and viral factors can predict the response of HCV-infected patients to IFN-α and ribavirin combination therapy. Studies showed that factors such as the HCV genotype 1b and viral load are associated with resistance of HCV-infected patients to INF therapy[37,38]. It was reported that resistance of HCV genotype 1b-infected patients to INF therapy is influenced by a region of the NS5A viral phenotype[23,39]. Mutations in this region, known as ISDR, are beneficial for patients receiving IFN therapy, whereas the wild type virus is resistant to IFN treatment[23,39]. Other studies fail to confirm the association between ISDR genotypes and IFN responsiveness[26,27,40-44], thus it remains a controversial issue[45,46].
Combined IFN and ribavirin therapy has replaced IFN monotherapy for HCV-infected patients about 10 years before. In the present study, to identify the predictive factors for effective combination therapy, we investigated the relationship between the response to combination therapy and a variety of factors. Only patients with HCV genotype 1b were studied because this genotype is known to be more resistant to interferon treatment than the other genotypes and is the most prevalent genotype in Korea[15,16].
In this study, age, PePHD mutations, HCV RNA titer, and ISDR subtype were found to be the predictive factors for combined IFN-α and ribavirin therapy for HCV genotype 1b infection. On the other hand, gender and ALT level were not associated with the combination therapy efficacy. These results are consistent with many previous studies, but contrary to others[26,27,31]. Such a difference indicates that these factors are not always accurate predictors for IFN response. This effect may be due to the pleiotropic nature of IFN activity, in addition to other cellular and viral genes that also modulate the effectiveness of INF therapy for chronic hepatitis C. This is the first study to determine the factors that predict the effectiveness of combination therapy in Korean patients. Therefore, this study may reflect the Korean genetic characteristics.
HCV seems to have a defense strategy against the host cellular responses induced by IFN[47]. The E2 protein appears to play a major role as a potential immune response target, and may interfere with cellular effectors induced by IFN[48]. Information about the clinical implications of E2 containing PePHD, is still limited. Analysis of a small series of HCV genotype 1-infected patients showed that amino acid sequence variability in the PePHD region was similar in responders and no-responders, indicating that the PePHD region is very stable over time[49-51]. In our study, a sequence analysis of the PePHD region in 50 patients found mutations in eight cases, all in the complete response group, suggesting that mutations in the PePHD region are associated with the response to combination therapy. In other studies, a few cases showed some PePHD mutations in the no-response group, though more mutations appeared in the complete response group[31,52]. Therefore, further study is needed to determine why mutations only occur in the complete response group of HCV-infected patients in Korea.
Some studies showed that the association of ISDR mutation rate with treatment response, but the other studies did not[31,45]. One of the Korean studies reported that the effect of INF monotherapy is not associated with the ISDR mutation rate[53]. It is not sure, but the different result may be due to the treatment methods and the sample size.
In conclusion, response of HCV genotype 1b-infected patients to combination therapy is influenced, at least in part, by HCV RNA titer, age, PePHD mutations, and ISDR subtype, but not by gender and ALT level. After adjusting for age and HCV RNA titer, ISDR subtype may be the most potent predictive factor for combination therapy efficacy in Korean chronic hepatitis patients with HCV genotype 1b.
The effectiveness of IFN-α and ribavirin combination therapy in Hepatitis C virus (HCV) infected patients is not satisfactory, especially in patients with HCV genotype 1b. IFN is also inconvenient to use, costly, and has a variety of possible complications. Therefore, it is necessary to determine the predictive factors for IFN therapy.
It is very important to know the pathogenesis of HCV-infected patients to block the progression of hepatocellular carcinoma.
This is the first study to determine the factors that predict the effectiveness of IFN-α and ribavirin combination therapy in Korean patients. In this study we used a large number of samples to determine the ISDR subtype, HCV RNA titer, age, and PePHD mutations.
The predictive factors for IFN-α and ribavirin combination therapy in patients with HCV genotype 1b can be used to select its candidates.
This article is of theoretical and practical importance. The results show that HCV RNA ISDR type may be an important factor for predicting the efficacy of IFN-α and ribavirin combination therapy in Korean patients.
S- Editor Zhu LH L- Editor Wang XL E- Editor Li HY
| 1. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [PubMed] |
| 2. | Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045-5055. [PubMed] |
| 3. | Rosenberg S. Recent advances in the molecular biology of hepatitis C virus. J Mol Biol. 2001;313:451-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Sharara AI, Hunt CM, Hamilton JD. Hepatitis C. Ann Intern Med. 1996;125:658-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 137] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 5. | Moussalli J, Opolon P, Poynard T. Management of hepatitis C. J Viral Hepat. 1998;5:73-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 6. | Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet. 1998;352:1426-1432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1667] [Cited by in RCA: 1640] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 7. | Lai MY, Kao JH, Yang PM, Wang JT, Chen PJ, Chan KW, Chu JS, Chen DS. Long-term efficacy of ribavirin plus interferon alfa in the treatment of chronic hepatitis C. Gastroenterology. 1996;111:1307-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 175] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2509] [Cited by in RCA: 2434] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 9. | Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 251] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 10. | Chang YJ, Byun KS. [Treatment of chronic hepatitis C]. Korean J Gastroenterol. 2004;44:301-307. [PubMed] |
| 11. | Patterson JL, Fernandez-Larsson R. Molecular mechanisms of action of ribavirin. Rev Infect Dis. 1990;12:1139-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 185] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | McHutchison JG, Poynard T. Combination therapy with interferon plus ribavirin for the initial treatment of chronic hepatitis C. Semin Liver Dis. 1999;19 Suppl 1:57-65. [PubMed] |
| 13. | Davis GL. New schedules of interferon for chronic hepatitis C. J Hepatol. 1999;31 Suppl 1:227-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Schalm SW, Weiland O, Hansen BE, Milella M, Lai MY, Hollander A, Michielsen PP, Bellobuono A, Chemello L, Pastore G. Interferon-ribavirin for chronic hepatitis C with and without cirrhosis: analysis of individual patient data of six controlled trials. Eurohep Study Group for Viral Hepatitis. Gastroenterology. 1999;117:408-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 113] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Lee DS, Sung YC, Whang YS. Distribution of HCV genotypes among blood donors, patients with chronic liver disease, hepatocellular carcinoma, and patients on maintenance hemodialysis in Korea. J Med Virol. 1996;49:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Yeh BI, Kim HW, Kim HS, Lee JY, Lee KH, Lee KM, Kim JS, Han KH. The prediction of interferon-alpha therapeutic effect by sequence variation of the HCV hypervariable region 1. Yonsei Med J. 1999;40:430-438. [PubMed] |
| 17. | Takada A, Tsutsumi M, Okanoue T, Matsushima T, Komatsu M, Fujiyama S. Distribution of the different subtypes of hepatitis C virus in Japan and the effects of interferon: a nationwide survey. J Gastroenterol Hepatol. 1996;11:201-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | McOmish F, Yap PL, Dow BC, Follett EA, Seed C, Keller AJ, Cobain TJ, Krusius T, Kolho E, Naukkarinen R. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol. 1994;32:884-892. [PubMed] |
| 19. | Nousbaum JB, Pol S, Nalpas B, Landais P, Berthelot P, Bréchot C. Hepatitis C virus type 1b (II) infection in France and Italy. Collaborative Study Group. Ann Intern Med. 1995;122:161-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 348] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 20. | Pagliaro L, Craxí A, Cammaá C, Tiné F, Di Marco V, Lo Iacono O, Almasio P. Interferon-alpha for chronic hepatitis C: an analysis of pretreatment clinical predictors of response. Hepatology. 1994;19:820-828. [PubMed] |
| 21. | Lin R, Liddle C, Byth K, Farrell GC. Virus and host factors are both important determinants of response to interferon treatment among patients with chronic hepatitis C. J Viral Hepat. 1996;3:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Shiratori Y, Kato N, Yokosuka O, Hashimoto E, Hayashi N, Nakamura A, Asada M, Kuroda H, Ohkubo H, Arakawa Y. Quantitative assays for hepatitis C virus in serum as predictors of the long-term response to interferon. J Hepatol. 1997;27:437-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 23. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 452] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 24. | Odeberg J, Yun Z, Sönnerborg A, Weiland O, Lundeberg J. Variation in the hepatitis C virus NS5a region in relation to hypervariable region 1 heterogeneity during interferon treatment. J Med Virol. 1998;56:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 25. | Puig-Basagoiti F, Sáiz JC, Forns X, Ampurdanès S, Giménez-Barcons M, Franco S, Sánchez-Fueyo A, Costa J, Sánchez-Tapias JM, Rodés J. Influence of the genetic heterogeneity of the ISDR and PePHD regions of hepatitis C virus on the response to interferon therapy in chronic hepatitis C. J Med Virol. 2001;65:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron JP, Lunel F, Stuyver L. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J Hepatol. 1997;27:72-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Squadrito G, Orlando ME, Cacciola I, Rumi MG, Artini M, Picciotto A, Loiacono O, Siciliano R, Levrero M, Raimondo G. Long-term response to interferon alpha is unrelated to "interferon sensitivity determining region" variability in patients with chronic hepatitis C virus-1b infection. J Hepatol. 1999;30:1023-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Chung RT, Monto A, Dienstag JL, Kaplan LM. Mutations in the NS5A region do not predict interferon-responsiveness in american patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 29. | Veillon P, Payan C, Gaudy C, Goudeau A, Lunel F. [Mutation analysis of ISDR and V3 domains of hepatitis C virus NS5A region before interferon therapy with or without ribavirin]. Pathol Biol (Paris). 2004;52:505-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Vuillermoz I, Khattab E, Sablon E, Ottevaere I, Durantel D, Vieux C, Trepo C, Zoulim F. Genetic variability of hepatitis C virus in chronically infected patients with viral breakthrough during interferon-ribavirin therapy. J Med Virol. 2004;74:41-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Hung CH, Lee CM, Lu SN, Lee JF, Wang JH, Tung HD, Chen TM, Hu TH, Chen WJ, Changchien CS. Mutations in the NS5A and E2-PePHD region of hepatitis C virus type 1b and correlation with the response to combination therapy with interferon and ribavirin. J Viral Hepat. 2003;10:87-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Murphy MD, Rosen HR, Marousek GI, Chou S. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig Dis Sci. 2002;47:1195-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Van der Poel CL, Cuypers HT, Reesink HW, Weiner AJ, Quan S, Di Nello R, Van Boven JJ, Winkel I, Mulder-Folkerts D, Exel-Oehlers PJ. Confirmation of hepatitis C virus infection by new four-antigen recombinant immunoblot assay. Lancet. 1991;337:317-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 365] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 34. | Han DP, Lee HW, Sohn JH, Yeh BI, Choi JW, Kim HW. The new genotypic human calicivirus isolated in Seoul. Exp Mol Med. 2000;32:6-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 35. | Sanger F, Coulson AR. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978;87:107-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 1188] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 36. | Choo SH, So HS, Cho JM, Ryu WS. Association of hepatitis C virus particles with immunoglobulin: a mechanism for persistent infection. J Gen Virol. 1995;76:2337-2341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J. Predictive factors of virological non-response to interferon-ribavirin combination therapy for patients infected with hepatitis C virus of genotype 1b and high viral load. J Med Virol. 2006;78:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 38. | Arase Y, Suzuki F, Tsubota A, Suzuki Y, Saitoh S, Kobayashi M, Akuta N, Someya T, Hosaka T, Kobayashi M. Sustained negativity for HCV-RNA over 24 or more months by long-term interferon therapy correlates with eradication of HCV in patients with hepatitis C virus genotype 1b and high viral load. Intervirology. 2004;47:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 39. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 734] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 40. | Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Bréchot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113:567-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 103] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 41. | Zeuzem S, Lee JH, Roth WK. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 42. | Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373-1381. [PubMed] |
| 43. | Rispeter K, Lu M, Zibert A, Wiese M, de Oliveira JM, Roggendorf M. The "interferon sensitivity determining region" of hepatitis C virus is a stable sequence element. J Hepatol. 1998;29:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 44. | Nakano I, Fukuda Y, Katano Y, Nakano S, Kumada T, Hayakawa T. Why is the interferon sensitivity-determining region (ISDR) system useful in Japan? J Hepatol. 1999;30:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Herion D, Hoofnagle JH. The interferon sensitivity determining region: all hepatitis C virus isolates are not the same. Hepatology. 1997;25:769-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Brechot C. The direct interplay between HCV NS5A protein and interferon transduction signal: from clinical to basic science. J Hepatol. 1999;30:1152-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 47. | He Y, Katze MG. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 2002;15:95-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 48. | Taylor DR. Hepatitis C virus: evasion of the interferon-induced antiviral response. J Mol Med (Berl). 2000;78:182-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 49. | Abid K, Quadri R, Negro F. Hepatitis C virus, the E2 envelope protein, and alpha-interferon resistance. Science. 2000;287:1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 50. | Gerotto M, Dal Pero F, Pontisso P, Noventa F, Gatta A, Alberti A. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology. 2000;119:1649-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 51. | Sarrazin C, Kornetzky I, Rüster B, Lee JH, Kronenberger B, Bruch K, Roth WK, Zeuzem S. Mutations within the E2 and NS5A protein in patients infected with hepatitis C virus type 3a and correlation with treatment response. Hepatology. 2000;31:1360-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 52. | Yang SS, Lai MY, Chen DS, Chen GH, Kao JH. Mutations in the NS5A and E2-PePHD regions of hepatitis C virus genotype 1b and response to combination therapy of interferon plus ribavirin. Liver Int. 2003;23:426-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 53. | Bae SH, Park YM, Yoo DG, Choi JY, Byun BH, Yang JM, Lee CD, Cha SB, Park DH, Kim BS. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence is not a predictive factor for response to interferon-alpha therapy and development of hepatocellular carcinoma. J Korean Med Sci. 2000;15:53-58. [PubMed] |