Published online Dec 14, 2007. doi: 10.3748/wjg.v13.i46.6166
Revised: September 15, 2007
Accepted: October 21, 2007
Published online: December 14, 2007
AIM: To identify the relationship between DNA hyper-methylation and histone modification at a hyperme-thylated, silenced tumor suppressor gene promoter in human gastric cancer cell lines and to elucidate whether alteration of DNA methylation could affect histone modification.
METHODS: We used chromatin immunoprecipitation (ChIP) assay to assess the status of histone acetylation and methylation in promoter regions of the p16 and mutL homolog 1 (MLH1) genes in 2 gastric cancer cell lines, SGC-7901 and MGC-803. We used methylation-specific PCR (MSP) to evaluate the effect of 5-Aza-2’-deoxycytidine (5-Aza-dC), trichostatin A (TSA) or their combination treatment on DNA methylation status. We used RT-PCR to determine whether alterations of histone modification status after 5-Aza-dC and TSA treatment are reflected in gene expression.
RESULTS: For the p16 and MLH1 genes in two cell lines, silenced loci associated with DNA hypermethylation were characterized by histone H3-K9 hypoacetylation and hypermethylation and histone H3-K4 hypomethylation. Treatment with TSA resulted in moderately increased histone H3-K9 acetylation at the silenced loci with no effect on histone H3-K9 methylation and minimal effects on gene expression. In contrast, treatment with 5-Aza-dC rapidly reduced histone H3-K9 methylation at the silenced loci and resulted in reactivation of the two genes. Combined treatment with 5-Aza-dC and TSA was synergistic in reactivating gene expression at the loci showing DNA hypermethylation. Similarly, histone H3-K4 methylation was not affected after TSA treatment, and increased moderately at the silenced loci after 5-Aza-dC treatment.
CONCLUSION: Hypermethylation of DNA in promoter CpG islands is related to transcriptional silencing of tumor suppressor genes. Histone H3-K9 methylation in different regions of the promoters studied correlates with DNA methylation status of each gene in gastric cancer cells. However, histone H3-K9 acetylation and H3-K4 methylation inversely correlate with DNA methylation status of each gene in gastric cancer cells. Alteration of DNA methylation affects histone modification.
- Citation: Meng CF, Zhu XJ, Peng G, Dai DQ. Re-expression of methylation-induced tumor suppressor gene silencing is associated with the state of histone modification in gastric cancer cell lines. World J Gastroenterol 2007; 13(46): 6166-6171
- URL: https://www.wjgnet.com/1007-9327/full/v13/i46/6166.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i46.6166
Multiple recent reviews have shown that virtually all human cancer types have epigenetic abnormalities that collaborate with genetic changes to drive cancer development and progression[1-7]. Hypermethylation of DNA in promoter CpG islands of tumor suppressor genes (TSGs) is known to inhibit transcriptional initiation and cause permanent silencing of the genes, which play a crucial role in carcinogenesis[1,2]. It was reported that hypermethylation of DNA in promoter CpG islands and diminished expression are present in a number of tumor-related genes in gastric cancer, which is one of the major current causes for cancer death in Asian countries[8]. For example, silencing of the cyclin-dependent kinase inhibitor p16 gene induced by hypermethylation can lead to disruption of cell cycle regulation and provide a growth advantage to affected cells[9]. A mismatch repair gene, MLH1, is often silenced with aberrant CpG island hypermethylation in gastric cancers[10,11]. Except for DNA methylation, recent studies have demonstrated the importance of histone modification as another epigenetic mechanism in the organization of chromosomal domains and gene regulation[12-18]. Acetylation of H3-K9 and methylation of H3-K4 are associated with open chromatin configurations such as that found at transcriptionally active promoters. In contrast, methylation of H3-K9 is a marker of condensed, inactive chromatin of the sort associated with the inactive X-chromosome and pericentromeric heterochromatin[16,17,19,20].
It has also been shown that histone modification is crucial to the process of DNA methylation in some organisms and abrogation of H3-K9 methylation in Neurospora results in loss of DNA methylation[21]. It was reported that histone H3-K9 methylation directly correlates with DNA methylation of some tumor suppressor genes, while histone H3-K9 acetylation and histone H3-K4 methylation inversely correlate with DNA methylation of some tumor suppressor genes[22]. These data suggest a functional linkage between DNA methylation and histone modifications in gene repression. To better understand the relationship between DNA methylation and histone modification in cancer-associated gene silencing, we performed ChIP assay to assess the methylation and acetylation of H3-K9 and the methylation of H3-K4 at the p16 and MLH1 genes in two gastric cancer cell lines. We also treated the gastric cancer cell lines with the DNA methylation inhibitor, 5-Aza-dC, and the histone deacetylase inhibitor, TSA, to elucidate whether alteration of DNA methylation affects histone modification.
Two cell lines derived from human gastric cancer, SGC-7901 and MGC-803, were cultured in RPMI 1640 supplemented with 10% fetal bovine serum(Gibco), penicillin (100 IU/mL) and streptomycin (100 μg/mL), and incubated in a humidified incubator containing 50 mL/L CO2 at 37°C.
TSA and 5-Aza-dC were purchased from Sigma. TSA was dissolved in absolute ethanol at a stock concentration of 3.3 mmol/L and stored at -80°C. 5-Aza-dC was dissolved in water at a stock concentration of 1 mmol/L and stored at -80°C. Cells were seeded at a low density in a 100 mm tissue culture dish and incubated for 24 h prior to treatment with 5-Aza-dC and TSA. 5-Aza-dC (5 μmol/L) was used for 72 h in the treatment. Culture medium containing 5-Aza-dC was exchanged every 24 h. TSA (300 nmol/L)was used for only 24 h in the treatment. 5-Aza-dC was used for 48 h followed by TSA for an additional 24 h in the combined treatment. Mock-treatment with an identical volume of absolute ethanol or water was used as a control.
The genomic DNA was modified by bisulfite treatment, as described previously[23]. DNA was purified using a Wizard DNA clean-up system (Promega), precipitated with ethanol, and resuspended in 30 μL of Tris-EDTA buffer. Two microliters of the aliquot was used as a template. The primers used for MSP and additional PCR conditions are described elsewhere[22]. PCR products were separated by electrophoresis on 2% agarose gels and quantitated with the FluorChem 2.0 system. The experiment was repeated three times.
Total cellular RNA was extracted from each of the two cell lines with TriZOL (Invitrogen) according to the manufacturer’s protocol. RNA was resuspended in nuclease-free water and quantitated with a spectrophotometer. Reverse transcription (RT) reactions were done on 2 μg of total RNA following the manufacturer’s protocol (Promega). cDNA was amplified by PCR using primers as described previously. Reaction conditions for each PCR are described elsewhere[24]. PCR products were resolved on 2% agarose gels and quantitated using the Fluor Chem 2.0 system. The level was determined by quantifying the intensities of the PCR product versus glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The experiment was repeated three times.
ChIP assays were performed as described previously with some modifications[24]. Briefly, proteins were cross-linked to DNA by adding formaldehyde directly into the culture medium to a final concentration of 4 g/L for 20 min at 37°C. After washing, the cell pellets were resuspended in 500 μL lysis buffer and sonicated thirty-five times, 2 s each. The average fragment size after sonication was 500 bp. The lysate (500 μL) was then divided into three fractions. The first and second fractions (200 μL each) were diluted in 1800 μL of lysis buffer, and the third fraction (100 μL) was used as an input control. The first lysate was incubated overnight at 4°C with 5 μL anti-Lys-9 acetylated histone H3 antibody, 5 μL anti-Lys-9 dimethylated histone H3 antibody, or 5 μL anti-Lys-4 dimethylated histone H3 antibody (all antibodies from Upstate Biotechnology) overnight at 4°C. The second lysate was incubated with Tris-EDTA buffer (5 μL) as a negative control. Immune complexes were collected with 20 μL protein A-sepharose beads for 1 h at 4°C with agitation. The cross-links were reversed by heating the sample at 65°C for 5 h. After elution, the samples were digested with proteinase K. DNA was recovered by phenol extraction, precipitated with ethanol, and resuspended in Tris-EDTA buffer.
Amplification was carried out with 2 μL of an immunoprecipitated DNA, a control without antibody or a 1:10 dilution of input DNA that was not immunoprecipitated. The primers used for ChIP and PCR conditions are described elsewhere[22]. We selected P16-3, P16-6, MLH1-2 and MLH1-3. PCR products were electrophoresed on 2% agarose gels and quantitated with the FluorChem 2.0 system. The level of histone acetylation and methylation in each immunoprecipitation was measured by quantifying the intensities of the PCR product in immunoprecipitated DNA versus input DNA diluted at 1:10 (total chromatin). The experiment was repeated three times.
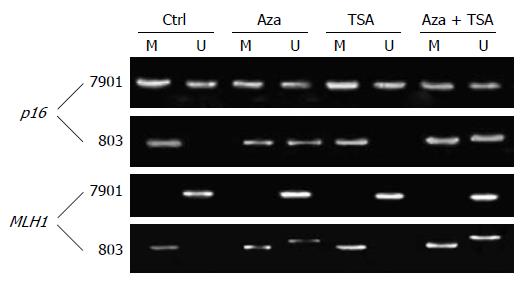
The two cell lines showed a characteristic DNA methy-lation status in each promoter region. As shown in Figure 1, p16 was hypermethylated (both alleles methylated) in MGC-803 and partially methylated (only one allele methylated) in SGC-7901. MLH1 was hypermethylated in MGC-803 but not methylated in SGC-7901.
5-Aza-dC and combined 5-Aza-dC and TSA resulted in demethylation of p16 and MLH1 in MGC-803, in which the silenced gene was associated with DNA hypermethylation. In contrast, TSA alone did not affect the DNA methylation status of p16 and MLH1.
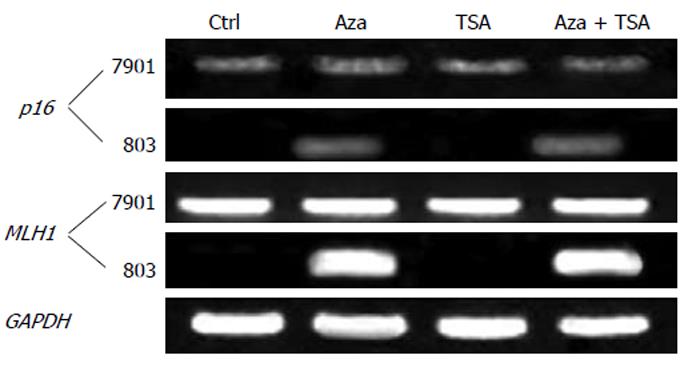
As shown in Figure 2, p16 was expressed in SGC-7901 and minimally affected by TSA. p16 was silenced in MGC-803 and TSA was not able to activate gene expression. In contrast, 5-Aza-dC alone reactivated expression of the p16 in MGC-803. Similar results were obtained in MLH1, which was expressed in SGC-7901 but silenced in MGC-803. TSA had no effect on gene expression, while 5-Aza-dC reactivated the silenced gene. The combined treatment with 5-Aza-dC and TSA increased gene expression.
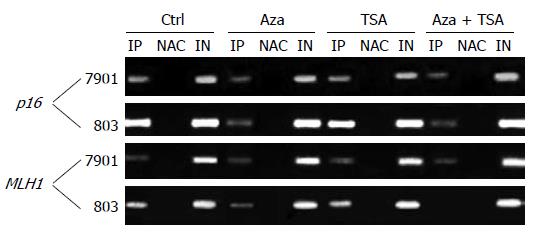
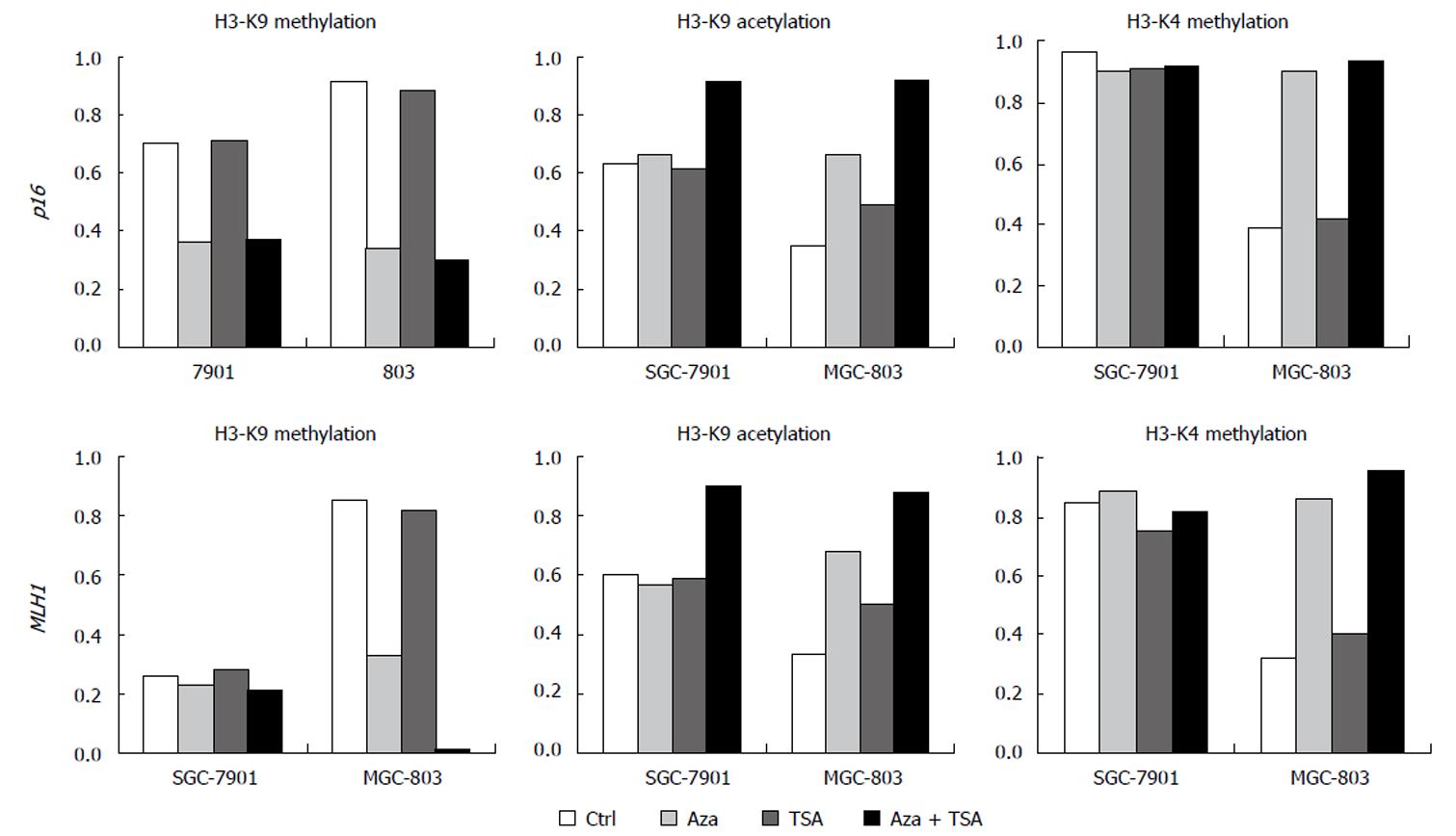
The results of ChIP studies were almost identical in different regions of each promoter, and the values for each gene were averaged to present the data. In the promoter region of the p16 gene, H3-K9 methylation was higher for MGC-803 than for SGC-7901. Similar results were seen at MLH1. SGC-7901 having no promoter DNA methylation at this locus, showed a low degree of H3-K9 methylation. MGC-803 had a higher degree of H3-K9 methylation across the promoter (Figures 3 and 4).
TSA alone had no effect on H3-K9 methylation, irrespective of DNA methylation status. In contrast, 5-Aza-dC had effects on H3-K9 methylation at the silenced loci, reducing histone H3-K9 methylation in the promoter showing partial methylation or hypermethylation (the promoter region of p16 in both cell lines and the promoter region of MLH1 in MGC-803). The combination of 5-Aza-dC and TSA had similar effects on histone H3-K9 methylation.
The promoter region of the p16 gene showed a higher degree of H3-K9 acetylation in SGC-7901 (partially methylated) than in MGC-803 (hypermethylated). Similar results were seen in the MLH1 gene showing a low degree of H3-K9 acetylation in all parts of the promoter region in MGC-803. In contrast, a higher degree of H3-K9 acetylation was detected in SGC-7901 (both alleles non-methylated) at all MLH1 regions studied (Figures 4 and 5A).
For the p16 gene, treatment with TSA alone had no effect on H3-K9 acetylation in the SGC-7901 (partial DNA methylation) but slightly increased H3-K9 acetylation in the silenced MGC-803. Identical results were seen in MLH1. 5-Aza-dC increased H3-K9 acetylation at the loci with DNA hypermethylation (p16 and MLH1 in MGC-803) but had no effect on the loci with partial or no DNA methylation (p16 and MLH1 in BGC-7901). However, the combination of 5-Aza-dC and TSA increased H3-K9 acetylation effectively at all loci irrespective of DNA methylation status. The results of ChIP studies were almost identical in different regions of each promoter.
For the p16 and MLH1 genes, H3-K4 methylation was higher in SGC-7901 than in MGC-803. TSA did not affect H3-K4 methylation. However, 5-Aza-dC, or the combination of 5-Aza-dC and TSA, increased H3-K4 methylation at all silenced loci in MGC-803. Little change in H3-K4 methylation was observed in SGC-7901, where only partial or no methylation was observed (Figures 4 and 5B).
Silencing of tumor suppressor genes is related to epigenetic regulation of both DNA methylation and histone modification[25]. In the present study, hypermethylation in promoter CpG islands was significantly associated with p16 and MLH1 silencing. Furthermore, aberrantly silenced and DNA hypermethylated genes in gastric cancer cells were characterized by histone H3-K9 hypermethylation, H3-K4 hypomethylation and H3-K9 hypoacetylation.
DNA methylation and histone modification may act synergistically or antagonistically on gene expression[26,27]. We carried out ChIP assays to explore the relationship between DNA methylation and histone modifications. ChIP is a powerful technique to test for the presence of certain DNA-binding proteins that might modulate chromatin structure and/or transcriptional characteristics of the specific region of DNA with which they are associated. We made use of polyclonal antibodies generated against methylated and acetylated histone H3, all of which are proteins linked to chromatin modification and regulation of transcription. In colorectal cancer, histone H3-K9 methylation directly correlates and histone H3-K4 methylation inversely correlates with DNA methylation of p16, MLH1 and the O6-methylguanine-DNA methyltransferase gene, MGMT[22]. We demonstrated that histone H3-K9 methylation correlated, and histone H3-K9 acetylation and H3-K4 methylation inversely correlated very well with DNA methylation of p16, MLH1 in SGC-7901 and MGC-803.
To further explore the relationship between DNA methylation and histone modification, we treated cancer cells with 5-Aza-dC and TSA. 5-Aza-dC, a DNA methyltransferase inhibitor, is sufficient to cause demethylation of the promoter region and reactivate expression of the hypermethylated, silenced gene[28,29]. TSA, a specific histone deacetylase (HDAC) inhibitor, has been permitted evaluation of the role of HDAC in silencing a variety of methylated genes[14]. It was reported that re-expression of DNA hypermethylated and silenced cancer genes can be induced through 5-Aza-dC-induced DNA demethylation, demethylated genes and the active marks, acetylated H3-K9 and methylated H3-K4 can be detected in HCT116 and DKO colon cancer[29-34]. However, one silencing mark, dimethylated H3-K9, is strikingly decreased[34]. In our study, when the CpG islands were hypermethylated, TSA increased histone acetylation, but had almost no effect on gene expression. In contrast, 5-Aza-dC reactivated expression of hypermethylation- induced silenced genes. Our findings on histone acetylation are consistent with previous reports linking the effect of DNA methylation and histone deacetylation on transcriptional silencing, demonstrating that DNA methylation is dominant over histone deacetylation in maintaining a silent state at hypermethylated promoters[22]. Furthermore, TSA and 5-Aza-dC play a different role in histone methylation. In the present study, 5-Aza-dC, but not TSA, could reactivate expression of the silenced genes and completely reverse key histone methylations surrounding the gene promoter, indicating that reactivation of silenced genes correlates much better with decreased histone H3-K9 methylation and increased H3-K4 methylation than with increased H3-K9 acetylation. We speculate that histone methylation plays a critical role in the maintenance of promoter DNA methylation-associated gene silencing in gastric cancer.
After 5-Aza-dC treatment, we observed a complete reversal of histone modification at the p16 and MLH1 promoter in MGC-803 cells. Acetylated H3-K9 and methyl-H3-K4 levels were increased, whereas methyl-H3-K9 levels decreased, suggesting that DNA hypermethylation may be essential for maintaining histone modification at gene promoters silenced due to aberrant DNA hypermethylation. DNA methylation plays a direct role in both genes silencing and maintaining a repressive histone modification at a hypermethylated gene promoter in cancer. Data show that DNMT1 interacts with HDAC activity in complexes bound to DNA, suggesting that it can recruit histone modifiers to DNA[33-35]. It was reported that DNA modification itself, or components of the DNA methylating machinery such as DNMTs or methyl-CpG binding proteins, can directly interact with histone methyltransferases or proteins in regions containing DNA methylation and allow them to set up an alterative histone modification[2], showing that histone methylation depends on DNA methylation.
Gastric cancer is a malignant tumor threatening human health worldwide. It has been shown that epigenetic mechanism plays an important role in the occurrence and development of gastric cancer. However, the role of histone modification and the relationship between DNA hypermethylation and histone modification at a hypermethylated, silenced tumor suppressor gene promoter in human gastric cancer remain unclear.
Histone modification plays a prominent role in the epigenetic regulation of gene transcription. There is evidence that dysregulation of epigenetic process causes transcriptional repression of a subset of genes, contributing to the pathogenesis of many cancers. The relationship between aberrant epigenetic changes and tumorigenesis is still to be identified.
In some cancers, histone H3-K9 methylation directly correlates and histone H3-K4 methylation inversely correlates with DNA methylation of some TSGs. Our findings may reflect the mechanism underlying inactivation of tumor suppressor genes by DNA hypermethylation and histone modification during gastric tumorigenesis. DNA methylation may affect histone modification in human gastric cancer.
Reactivation of the p16 and MLH1 gene by DNA methyltransferase inhibitor alone or in combination with histone deacetylase inhibitor suggests that the two agents can be used in treatment of gastric cancer.
Histone is the major component of chromatin functioning as a DNA packaging unit and as a transcriptional regulator. The amino-terminal tails of histones protrude from the nucleosome and are subjected to chemical modifications including phosphorylation, acetylation and methylation. These modifications of histones affect the access of regulatory factors and complexes to chromatin and influence gene expression. These different combinations of histone modifications at different residues may act synergistically or antagonistically on gene expression.
This is an important and interesting study, and the manuscript is well-written. The major finding in this paper is that treatment with 5-Aza-dC, a DNA methylation inhibitor and TSA, a histone deacetylase inhibitor, affects the expression of tumor suppressor genes, including p16 and MLH1, by reversing the hypermethylation and inhibiting histone deacetylation in human gastric cancer cells.
S- Editor Zhu LH L- Editor Wang XL E- Editor Ma WH
| 1. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. [PubMed] |
| 2. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1307] [Cited by in RCA: 1210] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 3. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1674] [Cited by in RCA: 1531] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 4. | Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315-2335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 340] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 5. | Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1182] [Cited by in RCA: 1185] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 6. | Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1264] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 7. | Zhu XJ, Dai DQ. Epigenetic modification and gastrointestinal tumor. Shijie Huaren Xiaohua Zazhi. 2006;14:3251-3256. |
| 8. | Fuchs CS, Mayer RJ. Gastric carcinoma. N Engl J Med. 1995;333:32-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 442] [Article Influence: 14.7] [Reference Citation Analysis (1)] |
| 9. | Shim YH, Kang GH, Ro JY. Correlation of p16 hypermethylation with p16 protein loss in sporadic gastric carcinomas. Lab Invest. 2000;80:689-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 98] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 10. | Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC. hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res. 1999;59:159-164. [PubMed] |
| 11. | Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090-1095. [PubMed] |
| 12. | Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453-2455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 464] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 13. | Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1307] [Cited by in RCA: 1301] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 14. | Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 838] [Cited by in RCA: 758] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 15. | Noma K CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150-1155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 560] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 16. | Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 369] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 17. | Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 275] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 18. | Xin Z, Allis CD, Wagstaff J. Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader-Willi syndrome imprinting center. Am J Hum Genet. 2001;69:1389-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Peters AH, Mermoud JE, O'Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 377] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 20. | Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 528] [Cited by in RCA: 520] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 21. | Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 743] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 22. | Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 263] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | Yang SH, Dai DQ. Analysis and improvement of methylation-specific polymerase chain reaction. Zhongliu Yanjiu Yu Linchuang. 2006;18:594-595. |
| 24. | Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein: DNA associations in a chromatin environment. Methods. 1999;19:425-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 463] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 25. | Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1449] [Cited by in RCA: 1394] [Article Influence: 53.6] [Reference Citation Analysis (0)] |
| 26. | Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7019] [Cited by in RCA: 6886] [Article Influence: 286.9] [Reference Citation Analysis (0)] |
| 27. | Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213-7218. [PubMed] |
| 28. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870-6875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1343] [Cited by in RCA: 1371] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 29. | Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456-6461. [PubMed] |
| 30. | Ghoshal K, Datta J, Majumder S, Bai S, Dong X, Parthun M, Jacob ST. Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure. Mol Cell Biol. 2002;22:8302-8319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 128] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | Guan ZY, Dai DQ, Meng CF. Experimental Studies on 5-aza-2'-deoxycitydine Induction for Demethylation and Up-regulated Transcription of Human Gastric Cancer Cell Lines TIMP3 Gene. Zhongguo Linchuang Zhongliu Zazhi. 2006;33:1334-1337. |
| 32. | McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541-3549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 213] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 33. | Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 691] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 34. | Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 761] [Cited by in RCA: 773] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 35. | Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 687] [Article Influence: 27.5] [Reference Citation Analysis (0)] |