INTRODUCTION
Colorectal cancer (CRC) is the second most prevalent cancer and the third leading cause of cancer death world-wide with almost 500000 related deaths every year[1]. Approximately half of all persons develop local recurrence or distant metastasis during the course of their illness, and the median survival time for these patients can vary from approximately 4 to 22 mo. The basis of treatment for metastasis or recurrent colorectal cancer is chemotherapy, although small number of patients can undergo surgery or others forms of loco regional treatment. While the Dukes and Tumor Node Metastasis (TNM) staging system identifies broad patients groups that vary in their long-term prognosis, considerable heterogeneity exists within each of different chemotherapy agents with regard to response to treatment.
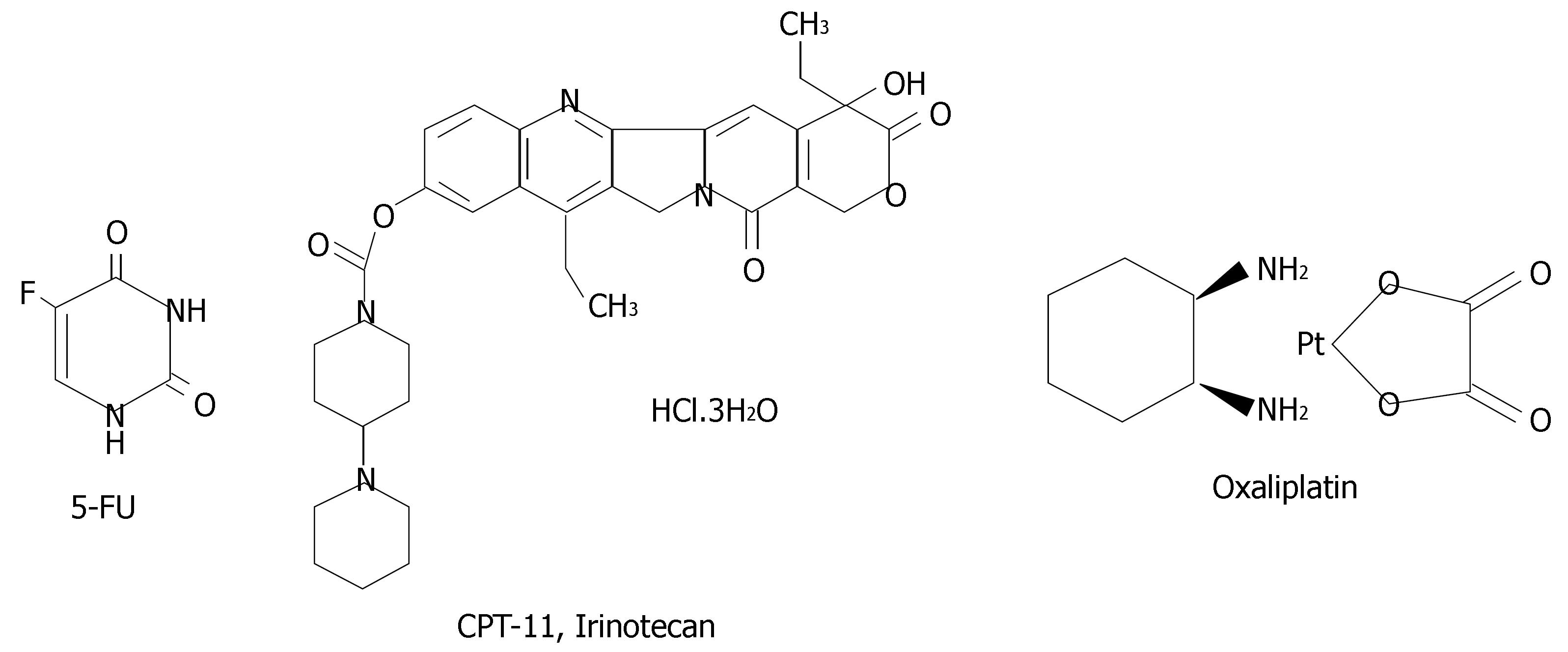
The most studied drug in CRC, the antimetabolite 5-fluorouracil (5-FU), was developed over 40 years ago. In the metastasis disease setting, single-agent 5-FU produced response rates of only 10%-20%[2]. Over the last 5 years, the median survival for patients with metastasis colorectal cancer has nearly doubled from 12-22 mo and the combination of 5-FU with new classes of drugs, such as oxaliplatin and CPT-11 (Irinotecan), has significantly improved response rates up into the 40%-50% range in patients with metastasis colorectal cancer[3]. Figure 1 shown chemical structure of these compounds. Furthermore, the use of novel biological agents, such as the monoclonal antibodies Cetuximab (an epidermal growth factor receptor (EGFR inhibitor) and Bevacizumab (a vascular endothelial growth factor (VEGF) inhibitor), have recently been shown to provide additional clinical benefit for patients with metastatic colorectal cancer[4,5].
Figure 1 Chemical structure of the three most important drugs used in colorectal chemotherapy: 5-FU, CPT11 and Oxaliplatin.
The objective of pharmacogenomics is to elucidate the complex genetic network responsible of drug efficacy and adverse drug reactions. The ultimate goal is to provide new strategies for optimizing the individual’s response to drug therapy based on patient’s genetic information[6]. Current methods of basing dosages on weight and age will be replaced with dosages based on an individual’s genetics. This will maximize the therapy’s value and decrease the likelihood of overdose.
In CRC, a limited number of predictive markers have been identified to date. The use of these as individual predictive markers has led to somewhat conflicting results. However, if these markers are used in combination they could provide a greater ability to reliably predict response to treatment[7]. Recent advances in our understanding of the molecular biology of CRC should lead to the identification of other panels of potential prognostic and predictive markers.
POLYMORPHISMS AND FLUOROPYRIMIDINES
To this day, the fluoropyrimidines (FPs) including 5-fluorouracil (5-FU), 5'-fluoro-2'-deoxyuridine, capecitabine, tegafur and S1, remain a major component of many standard regimens for numerous cancer types and a baseline component in many experimental regimens with novel agents[8]. Initially, 5-FU was the only effective systemic treatment for CRC, and since leucovorine enhances this effect, 5-FU and LV are given together[9]. FL reduces tumor size by 50% or more in approximately 20% of patients with advanced CRC, and prolongs median survival from approximately 6 mo to approximately 11 mo. When given as adjuvant therapy after the complete resection of tumor that has spread to regional lymph nodes (Stage III), FL increases the probability of remaining free of tumor at 5 years from approximately 42% to 58% and the likelihood of surviving for 5 years from 51% to 64%[10].
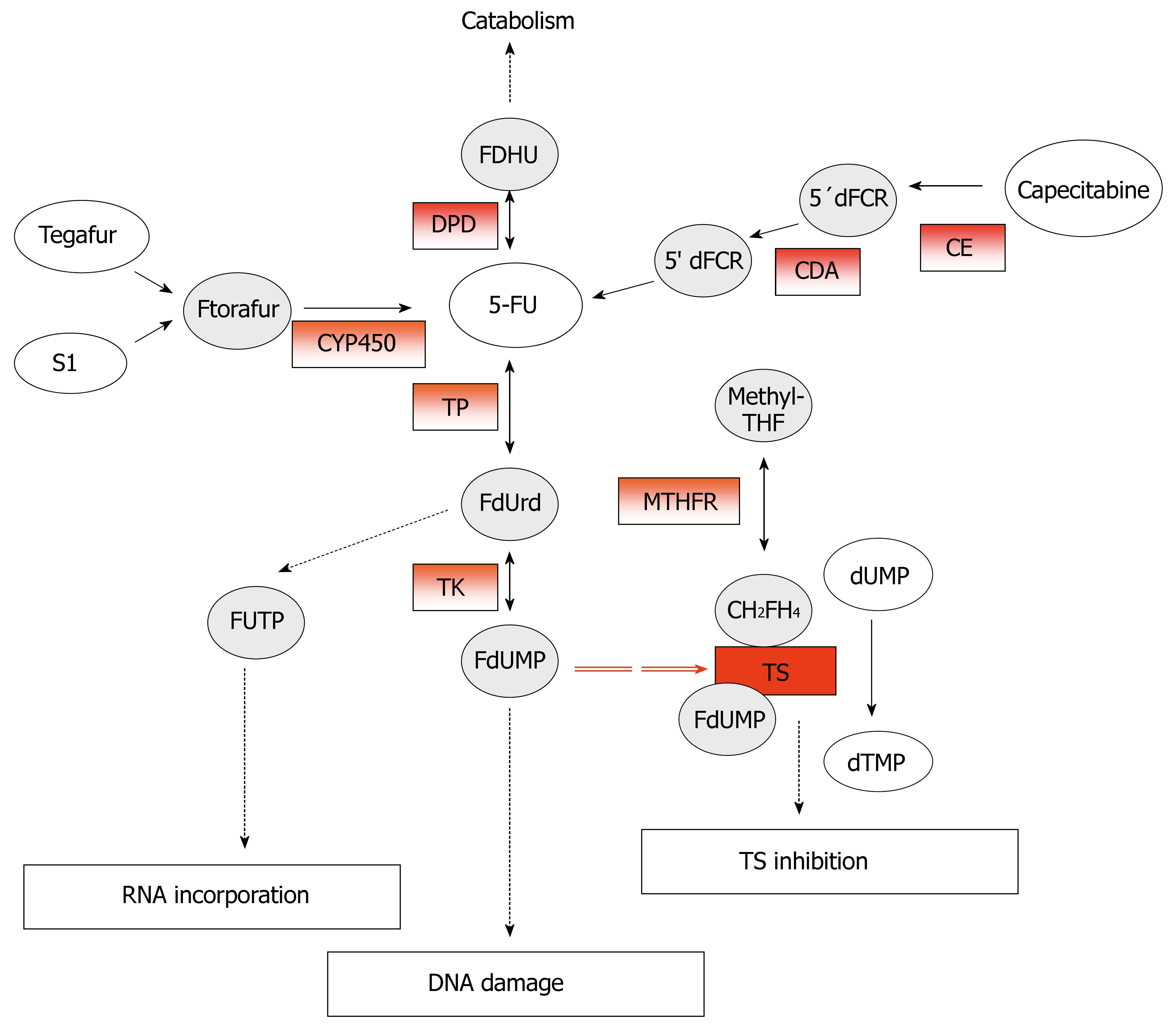
5-FU, an analog of uracil, is an anticancer prodrug that, after administration, is converted intracellular into three main active metabolites: 5-fluoro-2-deoxyuridine monophosphate (FdUMP), fluorodeoxyuridine triphosphate (FdUTP), and fluorouridine triphosphate (FUTP). The main toxic effects are mediated by the inhibition of thymidylate synthase (TS) through the formation of an extremely stable ternary complex among FdUMP, TS, and the cofactor 5, 10-methylene-tetrahydrofolate (CH2FH4)[11]. The formation of this complex prevents the methylation of the deoxyuridine -5'-monophosphate (dUMP) into deoxythymidine-5'-monophosphate (dTMP) catalyzed by TS. However, the incorporation of the FP metabolites, FdUTP and FUTP, into DNA and RNA respectively, contribute also to 5-FU cytotoxicity[12] (Figure 2).
Figure 2 Metabolism and mechanism of action of 5-fluoruracil (5-FU).
The potential predictive markers for 5-FU response are in red-boxes.
The common role played by FPs makes stratification according to likely response to this agent a relevant starting point in efforts to individualize treatment. For this purpose, reliable indicators for the prediction of the expected response are required. In the last few decades, intensive research aimed at understanding FP activity and extensive testing of patient’s outcomes have highlighted a number of characteristics as potential indicators of response.
Overexpression of TS has been reported in many types of tumors including breast, colon, gastric, and melanoma. In particular, TS overexpression has been found to be significantly associated with a low response to treatment based on 5-FU, both as adjuvant[13] and metastatic therapy[14] Several studies have proposed that genetic polymorphisms of TS gene can affect the response to 5-FU[15-17]. TS expression seems to depend on the number of the so-called TSER, tandem repeat polymorphic copies of 28 bp present in the 5'-promoter enhancer region of the gene[18]. TSER polymorphisms, therefore, are involved in the modulation of TS protein levels and can affect the drug response after administration of fluoropyrimidine. Most Caucasian subjects may be carriers of double (TSER*2) or triple (TSER*3) repetitions for this type of polymorphism, although there have also been reports of sequences with even more copies. An increase in the number of repeats gives rise to an increase in both mRNA and protein TS levels. Three copies of such repeats (TSER*3) lead to a TS expression which is 2.6 times higher than that produced by the presence of only two copies (TSER*2). Patients with CRCs, which show homozygote triple-tandem repeats (3R/3R), present high levels of intratumoral TS mRNA, elevated levels of TS protein, and a lower rate of response to chemotherapy than subjects with CRCs showing homozygote double-repeats (2R/2R)[19]. Similar results have been obtained in patients with metastatic CRCs[20]. Moreover, a study involving 221 Duke’s C stage CRC patients has shown that, with regard to survival rate, tumors with 3R/3R genotypes benefit less from chemotherapy than those with 2R/2R and 2R/3R genotypes[16]. A meta-analysis of 20 studies has made it possible to investigate the association between levels of TS expression and the survival of CRC patients[21]. The results have shown that high levels of TS in patients at any stage of the disease are predictive of outcome[22]. However, the predictive role of TS levels in early-stage CRC patients undergoing chemotherapy is still not fully understood; in fact, whereas in subjects undergoing surgery only, high TS levels are an independent prognostic factor for outcome, in those undergoing surgery and adjuvant FU, TS expression does not seem to predict outcome. Another study reports that in patients with advanced CRC treated with 5-FU/oxaliplatin, intratumoral TS levels appear to have an independent predictive value for survival[23]. Nevertheless, the data so far reported in literature are discordant; although, in fact, TS levels have prognostic value for CRC, this is lower in surgically-treated patients who undergo adjuvant therapy with 5-FU when the TS expression is low, but may be effective for tumors with high TS expression.
TP, also known as platelet-derived endothelial cell growth factor, catalyzes the conversion of 5-FU to the more active nucleoside form and has been shown to be an in vitro determinant of 5-FU activity. High expression of either TS or TP in colorectal tumors was shown to be an independent variable so that low expression of both enzymes in tumors predicted a very high expression rate to 5-FU as well as a significantly longer survival, whereas none of the patients with high expression of either TP or TS were responders. These data are in contrast to those demonstrating that cells with higher levels of TP should be more sensitive to 5-FU. These discrepancies may be due to the fact that high TP gene expression was not directly reflected in its protein products, and 5-FU metabolism may be limited by the availability of co substrates, or due to the role of TP as an angiogenic factor.
5-FU is inactivated in the liver by dihydropyrimidine dehydrogenase (DPD), which is the first key enzyme involved in the catabolism of the uracil and thymine into β-alanine. DPD activity is extremely variable in tumoral tissue and this variation might make a difference to the efficiency of 5-FU treatment, since intratumoral drug concentration is one of the most important factors for the determination of the antitumoral effect[24]. Deficiency in DPD activity, however, leads to severe toxicity correlated to 5-FU which may even be fatal. The partial or total lack of this enzyme has, in fact, been associated with severe toxicity (mucositis, granulocytopenia, and neuropathy), and in several cases even death, after 5-FU administration[25]. Analysis of the prevalence of various genetic variants of DPD among patients with DPD deficiency has shown that the most common mutation in DPYD is a G-A transition at the invariant GT splice donor site flanking exon 14 (IVS14 + 1G > A) in Caucasian populations; this mutation is responsible for the lack of exon 14 in mRNA transcript resulting in production of a truncated mRNA with virtually not present enzyme activity[26]. This allele is known as DPYD*2A and is one of the variants associated with severe toxicity after 5-FU treatment[27]. Recently two new missense mutations have been identified on codon 496 (A→G) in exon 6 and on codon 2846 (A→T) in exon 22, the latter in a patient with a total lack of DPD[28].
In the last few years, with the recognition that CH2FH4 was essential for the formation of the FdUMP-TS ternary complex, folate metabolism has also begin to emerge as a focus for FP response prediction. MTHFR converts CH2FH4 to 5-methyltetrahydrofolate. Consequently, it could be expected that the functionally comprised C677T variant would lead to increase CH2FH4 concentrations and thereby enhanced FP activity. Further support of a role for folate metabolism in determining FP response has been provided by the observation of a survival benefit from 5-FU treatment for colorectal cancer patients with DNA hypermethylation. Higher levels of folate intermediates, including CH2FH4, have been demonstrated in tumors with DNA hypermethylation[29]. Cohen and colleagues[30] found a statistically significant trend towards increased response to fluoropyrimidine-based chemotherapy with increasing copy number of the MTHFR 677 T allele in a study of 43 patients with metastatic colorectal cancer. In contrast, Wisotzkey and co-workers[31] did not observe a difference in survival by MTHFR C677T genotype among 51 Stage III colon cancer patients treated with 5-FU. However, both studies had a small number of subjects with the MTHFR 677TT genotype (n = 5), and lacked adjustment for potential confounding factors such as primary tumor site or type of chemotherapy received. Only one study has evaluated the effects of the MTHFR C677T, A1298C and TSER genotypes on time to progression and response to 5-FU-based treatment. Jakobsen and co-workers[32] studied 139 patients with metastatic colorectal cancer being treated in a randomized trial comparing three different 5-FU dosage levels. A greater percentage of individuals with the TSER 3R/3R or MTHFR 677T genotypes responded to treatment, and these same individuals had a statistically significant increase in time to disease progression for the first 8 mo post-treatment. However, later in the course there was no statistically significant difference in time to relapse by MTHFR or TS genotype.
Treatment of metastatic CRCs now includes the use of another chemotherapeutic agent, Capecitabine, which is an oral precursor of 5-FU. Due to its poor bioavailability and rapid catabolic clearance by DPD, 5-FU is unsuitable for oral delivery. Capecitabine or Xeloda® is a rationally designed oral fluoropyrimidine carbamate that, after selective conversion to 5-fluorouracil within solid tumors, acts by inhibiting thymidylate synthase activity. This would theoretically yield two advantages, enhanced drug concentrations at the tumor site and thus greater antitumor activity, and reduced drug levels in normal tissues with a consequent reduction in systemic toxicity.
Capecitabine is well absorbed by the gastrointestinal tract and undergoes a three-step enzymatic conversion to 5-FU. First metabolized in the liver by carboxylesterase to 5'-deoxy-5-fluorocytidine, capecitabine is converted in the liver and tumours tissues by citidine deaminase to 5'-deoxy-5-fluorouridine. A tumor-selective phenomenon is facilitated by higher intra-tumoral levels of thymidine-phosphorilase, the enzyme responsible for the final conversion step to 5-FU. With regard to 5-FU, low levels of TS and DPD lead to a better response to capecitabine. In particular, it has been observed that 75% of metastatic colorectal cancer patients, with homozygote double-repeat variants in TS (2R/2R), respond better to capecitabine administration compared with 8% of those with hetero-zygote variants (2R/3R) and 25% of those with triple-repeat homozygote variants (3R/3R)[33].
Recent advances in our understanding of the molecular biology of CRC should lead to the identification of other panels of potential prognostic and predictive markers associate with colorectal carcinogenesis.
In CRC, genetic instability has been recognized as a factor in the origin of malignant lesions, resulting in clonal evolution of genetic events acquired in the course of tumor progression. Microsatellite instability (MSI) is common to many forms of cancer and is found in 10%-14% of sporadic colon cancers[34]. MSI is caused by mutations in the mismatch repair (MMR) genes, such as hMSH2, hMLH1 and hMSH6, resulting in failure of the DNA MMR system to correct errors that occur during replication. An in vitro study[35] demonstrated that restoration of hMLH1 activity in the MMR-deficient HCT116 cells increased their sensitivity to 5-FU. Various studies have investigated the prognostic role of MSI in Stage II CRC. The studies have confirmed a consistent and independent association between MSI-high (MSI-H) phenotype and superior survival in Stage II and Stage III CRC patients[36]. Furthermore, Lim et al[37] demonstrated that patients with MSI tumors exhibited better recurrence-free survival compared with those with microsatellite stable (MSS) tumors. Moreover, the use of adjuvant chemotherapy did not benefit these patients. The use of MSI as a predictive marker of response to adjuvant chemotherapy still remains controversial. On the other hand, it has been reported that 70% of colorectal cancers have lost a portion of chromosome 17p, or 18q or both. The 17p chromosome contains the p53 gene, which is an important tumor suppressor, and is reported to be mutated in 40%-60% of colorectal cancers[38]. p53 status has been studied as a prognostic factor, and more recently as a predictor of response to cancer chemotherapy[39]. The study published by Tang and colleagues describe that p53 mutation was associated with a poorer prognosis in Stage II and III CRC patients who received surgery alone, whereas p53 was not a prognostic factor among those patients who had received 5-FU-based adjuvant chemotherapy[40]. However, Ahnen and co-workers found that patients with Stage III CRC, whose tumors overexpressed p53, did not derive significant survival benefit from adjuvant 5-FU-based treatment[41].
POLYMORPHISMS AND IRINOTECAN
The combination of 5-FU together with other drugs such as Irinotecan (CPT-11) has led to promising results in the treatment of CRCs, particularly in first line therapy of patients with metastatic disease. Partly as a result of the development of this agent, survival of patients suffering from incurable colorectal cancer has doubled during the last decade[42]. Like other camptothecins, the anti-neoplastic agent irinotecan (7-ethyl-10-[4-(1-piperidino)-1-piperidino]carbonyloxycamptothecin) and in particular its active metabolite SN-38 (7-ethyl-10-hydroxycamptothecin) stabilize the DNA-topoisomerase I complex by binding to it, preventing the resealing of single strand breaks[43]. Irinotecan prevents the replication division to proceed which results in double strand breaks and ultimately in its anti-tumor effect and its characteristic adverse effects on rapidly dividing tissues, such as bone marrow and intestinal mucosa. The main dose-limiting toxicities of irinotecan therapy are therefore myelosuppression and delayed-type diarrhea[44,45].
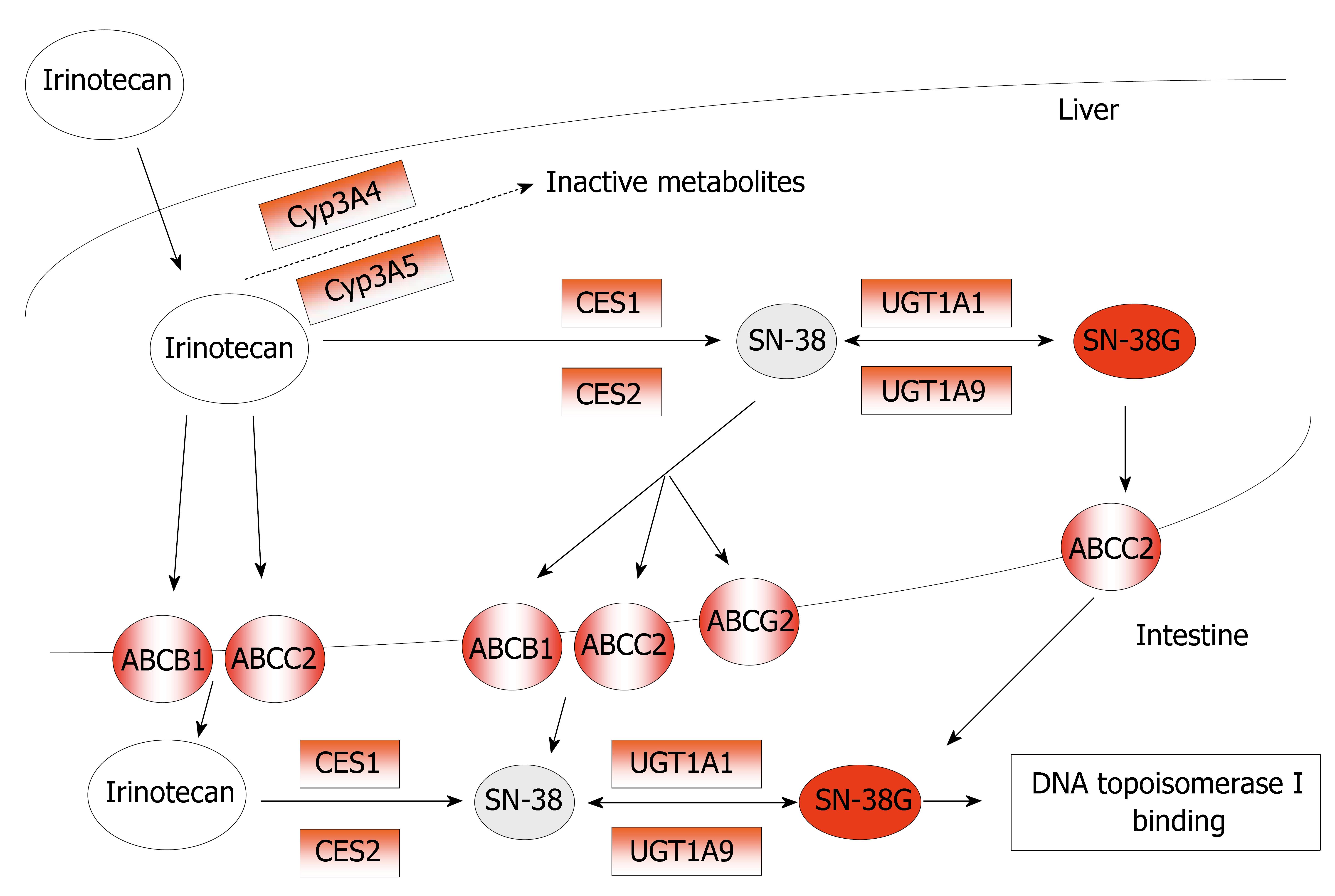
In humans, irinotecan is hydrolyzed into its active metabolite SN-38 by carboxylesterases, present in serum, intestines, tumor tissue, and in high content in the liver[46]. Recently, the opinion is emerging that intra-tumoral activation of irinotecan into SN-38 by CES might be even more important than systemic circulating SN-38 levels, formed by hepatic CES[47]. Although plasma levels of SN-38 are relatively low, relations between SN-38 and myelosuppression and/or diarrhea have been demonstrated[48]. Uridine diphosphate-glucurono-syltransferase 1A (UGT1A) mediated glucuronidation of SN-38, forming a β-glucuronic acid conjugate (SN-38G; 10-O-glucuronyl-SN-38), is the main pathway of detoxification for SN-38. Irinotecan is also sensitive to cytochrome P450 3A (CYP3A) that mediated oxidative pathways, resulting in the formation of inactive metabolites. Moreover, irinotecan, SN-38, and their metabolites are excreted by drug-transporting proteins from the adenosine-triphosphate binding cassette (ABC) transporter superfamily[49] (Figure 3).
Figure 3 Metabolism and mechanism of action of Irinotecan (CPT-11).
The potential predictive markers for CPT-11 response are in red-boxes.
The CES genes, located on chromosome 16q13-q22, are supposed to be highly conserved during evolution. However, recently, several polymorphisms in the CES-genes have been described, some of which with major racial differences in distribution[50]. Although the interpatient variation in CES activity is high and some SNPs appear to be very common[51], the functional consequences of reported SNPs on the in vivo activation of irinotecan into SN-38 are thought to be limited. Marsh et al[50] did not demonstrate any functional relationship between the presence of SNPs in the CES genes and CES mRNA levels, except for an intronic SNP (IVS10-88) in CES2 which was associated with reduced CES2 mRNA expression in colorectal tumors, but not in normal colonic mucosa. Neither did Charasson et al[52] find any influence of 11 silent SNPs in CES2 on gene expression or functional activity. Lack of association may be explained by the ineffective activation of irinotecan by CES, the role of other esterases, and the complex metabolic pathway of irinotecan. It may also be possible those other proteins regulate CES transcription and translation, or that other factors are rate limiting in the formation of active CES. However, as SNPs in CES may lead to less transcription and thus might lead to diminished local activation of irinotecan and less favorable therapeutic responses, both in vitro and in vivo functional investigation of SNPs in the CES genes is needed, especially of recently discovered SNPs in CES2.
Members of the cytochrome P450 superfamily are capable to oxidize more than half of all anti-cancer drugs. Especially the CYP3A subfamily, and in particular, the genes CYP3A4, CYP3A5, CYP3A7, and CYP3A43 are the most important. CYP3A4*1B, a SNP in the promoter area of the gene, was thought to be a promising polymorphism for irinotecan pharmacokinetics, partly as a result of its relatively high allele frequency compared to most other CYP3A4 SNPs[53]. However, Garcia-Martin et al[54] reported that the presence of CYP3A4*1B did not correlate with low enzyme activity in Caucasians. In a polygenetic approach to assess genotypes from multiple irinotecan pathway genes with irinotecan pharmacokinetics no effect on irinotecan pharmacokinetics was seen, neither for this SNP nor for the other studied CYP3A SNPs (CYP3A4*2, CYP3A4*3, CYP3A5*3 and CYP3A5*6)[55].
The human UGT superfamily has been classified into the UGT1 and UGT2 families, further classified into three subfamilies (UGT1A, UGT2A, and UGT2B)[56]. All nine functional members of the UGT1A subfamily are encoded by a single gene locus, the UGT1A locus on chromosome 2q37. Especially the UGT isoforms 1A1, 1A7 and 1A9 are involved in the phase II conjugation of SN-38 to the inactive metabolite SN-38G[57]. UGT1A1 and UGT1A9 are highly expressed in the gastrointestinal tract and the liver; the primary organ involved in the detoxification of irinotecan. Polymorphisms, resulting in absent or very low UGT1A1 activity, have been associated with three heritable unconjugated hyperbilirubinemia syndromes: Crigler-Najjar syndrome type 1 and 2[58], and Gilbert’s syndrome[59]. Gilbert’s syndrome is common among Caucasians and is associated with the presence of an extra, seventh, dinucleotide (TA) insertion (UGT1A1*28) in the (TA)6TAA-box of the UGT1A1 promoter region, leading to a considerable reduced enzyme expression of about 30%-80%. The UGT1A1 activity appears to be inversely related to the number of TA-repeats, varying from 5 to 8.
Studies have shown that the homozygous UGT1A1*28 genotype was associated with an increased risk of developing leucopenia and severe delayed-type diarrhea after treatment with irinotecan. Ando et al[60] analyze the association between UGT1A1 variants and irinotecan toxicity, revealing in a multivariate analysis that presence of UGT1A1*28 allele was a risk factor for severe toxicity. These data have been confirmed by other groups[9,61]. Based on this knowledge and the finding that demonstrated a good concordance between the UGT1A1*28 genotype and less effective SN-38 glucuronidation prospective studies were initiated. A significant relation was observed between the AUC of SN-38 and the number of TA-alleles[62]. In addition, two other promoter variants (UGT1A1-3279G>T and UGT1A1-3156G>A) have been identified. These variants are in strong linkage disequilibrium with the UGT1A1*28 polymorphism in Caucasians, while this link is less apparent in African-Americans and Asians, suggesting a different haplotype structure among various races[63]. Ando et al[64] found a strong a relation for presence of the UGT1A1-3263T>G SNP and the severity of irinotecan induced toxicity, although in a multivariate analysis including UGT1A1*28 as well, this effect was mainly attributed to this latter polymorphism[65]. Presented observations clearly illustrate that UGT1A1 mutations can influence a patient’s exposure to SN-38, and, hence, the susceptibility to toxicity. Recently, a study in colorectal cancer cell lines shown that DNA methylation represses UGT1A1 expression and that this process may contribute to the level of tumoral inactivation of the anticancer agent SN38 and potentially influence in clinical response[66].
The adenosine-triphosphate (ATP) binding cassette (ABC) transporters are the largest family of transmembrane proteins that use ATP-derived energy to transport various substances over cell membranes[67]. Their localization pattern suggests that they have an important role in the prevention of absorption and the excretion of potentially toxic metabolites and xenobiotics, including irinotecan and its metabolites.
P-glycoprotein, located on chromosome 7q21, and, among others, expressed in kidney, liver, and intestine, is known for more than 50 SNPs and other polymorphisms in the gene encoding this transporter[68]. Three SNPs which show linkage disequilibrium (ABCB1 1236C>T, ABCB1 2677G>A/T, and ABCB1 3435C>T), have been studied extensively [69]. However, a relation with irinotecan or its metabolites has been not demonstrated in Caucasians. Recently, Balram et al[70] showed a relation for ABCB1 3435C>T with irinotecan AUC (area under concentration versus time curves) in a small Chinese population which may be the result of lowered pump activity. In a group of 46 Caucasian patients, a significant effect of the ABCB1 1236C>T polymorphism on the AUCs of irinotecan and SN-38 was seen, resulting in an increase in both AUCs[71]. Although an effect of these three related SNPs on irinotecan pharmacokinetics seems likely, the true clinical relevance of their effects still remains to be clarified.
For the canalicular multispecific organic anion transporter (ABCC2), recently a functional SNP in irinotecan pharmacokinetics has been found (ABCC2 3972C>T). This SNP, studied in 64 Caucasian patients, resulted in highly significant effects on the AUC of irinotecan, and SN-38G, all being higher in patients carrying two 3972T alleles.
In vitro studies have indicated that the irinotecan metabolites SN-38 and its glucuronide conjugate SN-38G are very good substrates for the breast cancer resistance protein[72]. ABCG2, located on chromosome 4q22, was first found to be overexpressed in cancer cells with acquired resistance to anticancer drugs[73]. The ABCG2 gene is supposed to be well conserved and most SNPs found up to now seem unlikely to alter transporter stability or function[74]. Few SNPs with presumed clinical consequence have been studied in relation to irinotecan pharmacokinetics; in particular, a single-nucleotide polymorphism in exon 5 has been described. This ABCG2 421C>A transversion results in an amino acid change of glutamine to lysine at codon 141[75]. Functional consequences of this SNP were demonstrated in Caucasian cancer patients treated with the structurally related camptothecins diflomotecan and topotecan[76]. Patients carrying at least one defective ABCG2 421A allele were found to have higher drug levels. However, in a large group of Caucasian patients pharmacokinetic parameters of irinotecan and SN-38 were not significantly different[77].
POLYMORPHISMS AND OXALIPLATIN
Oxaliplatin (OXA), a third-generation platinum analog that distorts DNA adducts, administered alone or in combination with 5-FU/LV has broaden the therapeutic choices for patients with advanced CRC who may experience hepatic and pulmonary metastasis. The cytotoxic activity of oxaliplatin is initiated by formation of a DNA adduct between the adequated oxaliplatin derivative and a DNA base[78]. Initially, only monoadducts are formed but eventually oxaliplatin attaches simultaneously to two different nucleotide bases resulting in DNA cross-links. The adducts are formed with the N-7 positions of guanine and adenine preferentially and in most cases these reactions result in intrastrand cross-links. In the cell approximately one of every 100000 bases can be cross-linked by a platinum atom, resulting in 10000 platinum atoms per cell[79].
In general, the cytotoxic efficacy of platinum compounds in cancer cells can be related to inhibition of DNA synthesis or to saturation of the cellular capacity to repair Pt-DNA adducts. Platinum atoms modify the three-dimensional DNA structure, which inhibits the normal DNA synthesis and repair processes[80].
Interestingly, cellular DNA repair mechanisms seem to differ in their response to Pt or Pt-DACH complexes. After DNA-adduct formation by oxaliplatin, cells will activate cellular repair mechanisms. In general, DNA repair is carried out by specific enzymes that consist of several amino- and sulphur groups. Therefore, oxaliplatin can be covalently bound to these repair enzymes as well, impairing their function[81]. If substantial DNA damage persists this may ultimately lead to the activation of apoptotic pathways and cell death[82].
Several mechanisms are described that confer resistance to oxaliplatin, including diminished cellular drug accumulation, increased intracellular drug detoxification and increased Pt-DNA adduct repair. However, the overall sensitivity of a cell is multifactorial and the relative importance of each process on ultimate drug sensitivity is difficult to predict[83]. There is growing evidence that common gene variants affect the activity of cellular DNA repair and platinum conjugation.
The uptake of platinum by cells is not completely understood but there is evidence that decreased accu-mulation is the most common mechanism of resistance to cisplatin[82]. Platinum uptake by cells is an energy requiring process, but it is not saturable and possibly involves transport by a yet unidentified efflux pump. Once inside the cell, conjugation to glutathione (catalyzed by the enzyme glutathione-S-transferase, GST) effectively inactivates platinum compounds before DNA damage is induced. This conjugation reaction is followed by cellular excretion and is therefore related to cellular drug resistance as well. A number of studies indicate an important role of GST in oxaliplatin resistance. A single nucleotide polymorphism (SNP) in exon 5 at position 313 (A→G) in the GSTP1 (π) gene results causes the amino acid change Ile105→Val. The mutant GSTP1 (π) enzyme is less potent in detoxification of carcinogens and individuals with two mutant alleles have shown a significant survival benefit from combined oxaliplatin/5-FU treatment[84]. Other common polymorphisms in the GSTT1 (theta;) and GSTM1 (μ) genes include deletions that result in complete loss of enzyme activity in homozygous individuals. However, no association with altered survival or clinical response in patients with advanced colorectal cancer treated with oxaliplatin/5-FU was observed for the GSTT1 and GSTM1 genotypes[85].
Since the primary anti-tumor mechanism of oxaliplatin is the formation of Pt-DNA adducts, polymorphisms in genes involving the repair of these adducts, such as nucleotide excision repair, base excision repair, mismatch repair (MMR) and other post-replicative repair pathways, may affect oxaliplatin efficacy. Induction of the enzymes involved in these systems results in increased DNA repair activity, more efficient adduct removal and hence decreased sensitivity to platinum drugs.
Mismatch repair (MMR) is a DNA repair pathway that corrects base mispairs and small strand loops that occur during replication. Loss of MMR function results in an increased spontaneous mutation rate. The MMR system consists of six different proteins, originating from the hMLH1, hMLH2, hPMS2, hMSH2, hMSH3 and hMSH6 genes. In vitro studies showed that MMR is not involved in oxaliplatin induced DNA-damage repair, whereas it serves as an important mechanism in cisplatin and carboplatin adduct repair[86]. The conformational distortion of the oxaliplatin DNA complex is different from the cisplatin and carboplatin adduct and this, together with the less polar properties of the DACH-ligand, contributes to a recognition failure of MMR proteins to detect oxaliplatin adducts. To date, no polymorphisms in the MMR pathway genes are known that influence the anti-tumor effects of oxaliplatin.
Single-strand breaks resulting from exposure to endogenously produced active oxygen, ionizing radiation or alkylating agents are repaired by the base excision repair system. X-ray repair cross-complementing group 1 enzyme (XRCC1) contains a domain which functions as a protein-protein interface that interacts with poly (ADP-ribose) polymerase (PARP). Shen et al[87] identified three polymorphisms in the XRCC1 gene. One of these, located in exon 10 of this gene, causes the amino acid change Arg399→Gln in the PARP binding domain. The polymorphic enzyme is supposed to be less capable of initiating DNA repair due to altered binding characteristics. In individuals with the mutant Arg399→Gln codon increased DNA damage marker levels are found due to inadequate repair or increased damage tolerance. Patients with at least one of the mutant alleles have a more than five old risk of combined oxaliplatin/5-FU chemotherapy failure compared to patients with two wild type alleles[88].
Nucleotide excision repair is a pathway involved in the recognition and repair of damaged or inappropriate nucleotides. A wide variety of DNA-damage is repaired by NER, including UV-induced photo-products, helix-distorting monoadducts, cross-links and endogenous oxidative damage. At least six proteins are essential for damage recognition and removal by this repair pathway. The first step in this process is recognition of a damaged or inappropriate base by XPA (xeroderma pigmentosum complementation group A protein) and RPA (replication protein A). The adhesion of XPA and RPA to a DNA strand attracts other repair factors to the site followed by enzymatic unwinding of the helix lesion area by XPD. The XPD gene, also known as ERCC2 (excision repair cross complementing group 2), encodes an ATP-dependent helicase that is a component of transcription factor TFIIH. A significant relationship with clinical response to platinum-based chemotherapy was found for the Lys751→Gln polymorphism of ERCC2[89]. This SNP causes an amino acid change in exon 23 and apparently affecting protein function but not resulting in an alteration of any of the seven helicase domains. Metastatic colorectal cancer patients treated with oxaliplatin/5-FU showed different tumor response for the various genotypes; 24% responders in the Lys/Lys group, versus 10% in the Lys/Gln and 10% in the Gln/Gln groups, respectively[90]. Nevertheless, further studies are necessary in order to confim these data and to establish the real importance of polymorphisms in the gene XPD with regard to resistance to platinum agents.
TARGETED-THERAPIES FOR COLORECTAL CANCER
Targeted therapy is defined as a treatment with a focused mechanism that specifically acts on a well-defined target or biological pathway. The ideal cancer target can be defined as a macromolecule that is crucial to the malignant phenotype and is not expressed significantly in vital organs and tissues bind to cancer cells with high affinity and create anti-tumor effects.
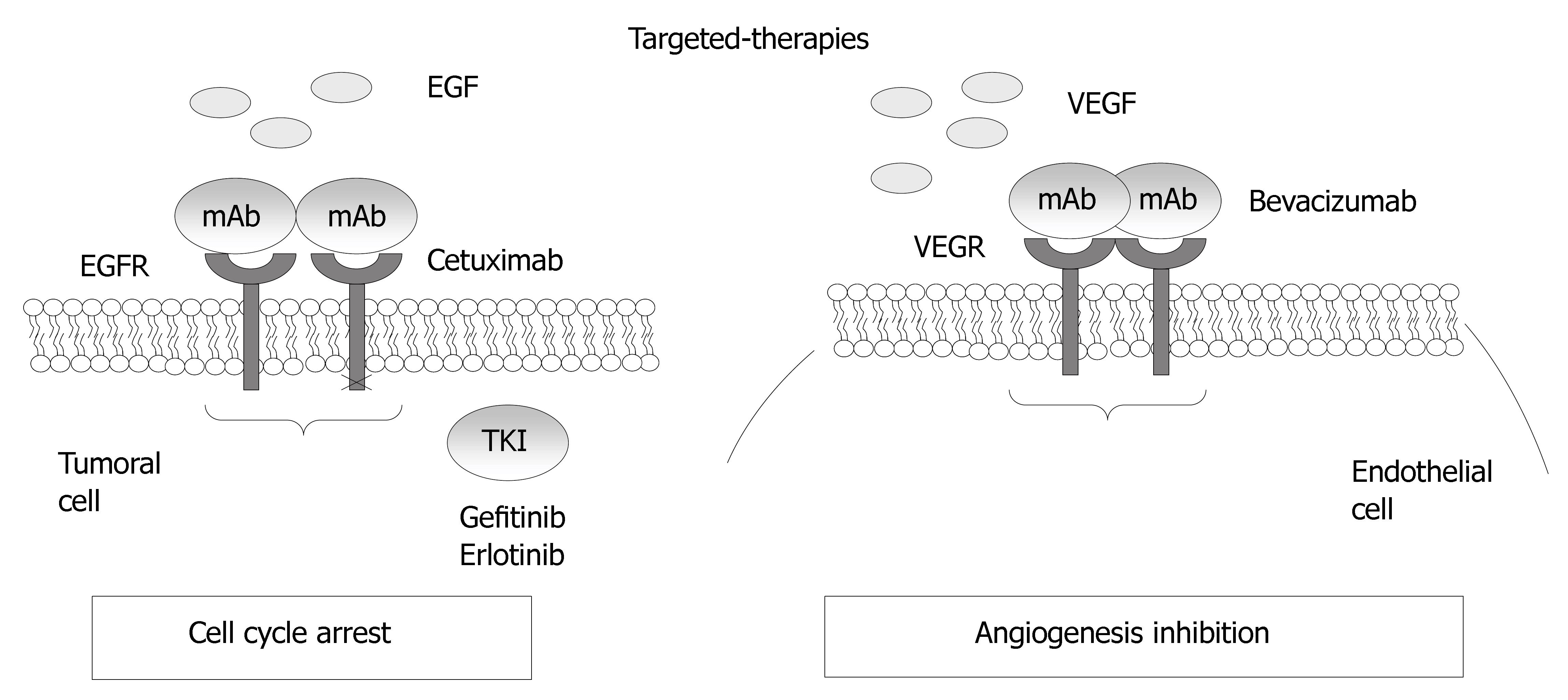
In colorectal cancer, two targets, the process of angiogenesis, and the epidermal growth factor receptor, are exploited by the newest monoclonal antibodies that are available for use in CRC patients (Figure 4).
Figure 4 Mechanisms of action for the epidermal growth factor receptor and VEGF.
EGFR-based therapies
EGFR is a tyrosine kinase receptor of the ErbB family that is abnormally activated in epithelial tumors, including 25%-80% of CRCs[91]. EGFR is a 170-kDa cell surface glycoprotein containing three well-identified parts: an extracellular binding domain, a hydrophobic membrane-spanning domain and a cytoplasmic domain containing the tyrosine kinase activity. The bind of specific ligands, EGF and TGFα, to the extracellular domain, leading to dimerization of the receptor with another EGFR (homodimerization) or another member of the EGFR family (heterodimerization). Its activation leads to downstream signaling that stimulates mitogenic and survival pathways such as mitogen-activated protein kinases (MAPKs) and phosphotidylinositol-3 kinase (PI3K)/Akt, which have tumor-promoting activities. Inhibition of these signaling pathways by EGFR antagonists can lead to induction of Bax, activation of caspase-8 and downregulation of Bcl-2 and NF-κB, initiating a cascade of intracellular signaling that ultimately regulates cell proliferation, migration, adhesion, differentiation, and survival[92,93]. Tumor cells that may be activated by ligands such as EGFR and TGFα may then become chemosensitive through EGFR inhibition and activation of these apoptotic pathways.
Agents targeted against the EGFR have been studied extensively in the laboratory, and several have undergone clinical trials, including Cetuximab (Erbitux), a humanized monoclonal antibody directed against the extracellular domain of the EGFR, and the small molecule tyrosine kinase inhibitors (TKIs) Gefitinib (Iressa/ZD1839), and Erlotinib (Tarceva/OSI-774).
Cetuximab binds to the EGFR with high affinity, blocking growth-factor binding, receptor activation, and subsequent signal-transduction events[94]. Preclinical models demonstrated modest in vitro and in vivo single-agent activity of Cetuximab but significant enhancing activity in combination with cytotoxic chemotherapy[95]. Cetuximab enhanced the antitumor effects of chemotherapy and radiotherapy by inhibiting cell proliferation, angiogenesis, and metastasis and by promoting apoptosis[92]. Several studies have shown that cetuximab is effective in patients with metastasic CRC whose disease has progressed on irinotecan-based chemotherapy. A phase II study of cetuximab monotherapy in EGFR-positive advanced CRC patients that failed a previous treatment with irinotecan, obtained 10.5% partial responses and disease stabilization in 35% patients[96]. The result of a multicenter phase II study in 246 advanced CRC patients that failed two lines of chemotherapy containing fluoropyrimidines, oxaliplatin and irinotecan have confirmed a partial response of 12% and a disease stabilization rate of 34%. The most important data for the use of cetuximabb, was derived from a large European randomized study, the BOND study, which compared cetuximab with cetuximab in association with irinotecan. Partial response were obtained in 22.9% patients treated with irinotecan plus cetuximab and the time of disease control was 55.5 mo[4].
The development of cetuximab in colorectal cancer was grounded on the premise that EGFR expression by IHC would be prognostic for cetuximab activity, with all trials to date requiring EGFR positivity by IHC. However, Chung et al[97] demonstrate no correlation between intensity of EGFR expression and clinical response, challenging this premise. The BOND study results, obtained similar conclusion and the probability or achieving a response was not correlated to the level of EGFR expression in the tumor[4]. On this basis, EGFR-negative colorectal cancer patients would not be excluded from standard protocol treatment with cetuximab on the basis of EGFR status. EGFR analysis by current IHC techniques does not appear to have predictive value, and selection or exclusion of patients for cetuximab therapy on the basis of currently available EGFR IHC does not appear reasonable[98]. This may be due in part to the lack of a standardized protocol and grading system for EGFR expression in clinical samples to technical limitations that are inherent in immunohistochemical methods or, perhaps, to an intrinsically poor correlation between the level of EGFR expression and therapeutic response.
A polymorphic (CA)n dinucleotide repeat is observed in intron 1 of the EGFR gene, which has been shown to be associated with gene expression[99]. It has been demonstrated that as the number of (CA)n repeats increases the level of transcription decreases[100]. However, in CRC cancer, association between the repeat length and EGFR protein expression was not been reported[101]. Neither, polymorphisms of EGFR has been associated with cetuximab therapy.
In addition to cetuximab, several tyrosine kinase inhibitors have been developed to target EGFR. A recent phase II study shown that the combination of capecitabine, oxaliplatin, and erlotinib seems to have promising activity against metastatic colorectal cancer in patients who received prior chemotherapy, with a relatively higher response rate and progression-free survival compared with previous reports of either infusional FU, leucovorin, and oxaliplatin or capecitabine and oxaliplatin in similar patient populations[102].
Skin rash has been the most commonly observed toxicity associated with the various EGFR inhibitors; interindividual differences in the onset, duration and severity of the rash have been observed, and no threshold plasma levels have been linked to the occurrence of the rash. Most intriguing are emerging data demonstrating a significant correlation between skin rash and survival among various patients treated with different anti-EGFR therapies. There are several potential hypotheses being put forward to explain both the variable toxicity and efficacy of EGFR inhibitors. One such hypothesis proposes that variability in clinical observations is related to variable drug exposure. For example, the small-molecule EGFR tyrosine kinase inhibitors gefitinib and erlotinib are metabolized by CYP3A, and it is certainly plausible that individuals with variant CYP3A alleles might have differences in drug exposure. On the other hand, the previously described CA dinucleotide repeat polymorphism might influence the drug response due to differences in target expression. Data that indirectly lend support to this hypothesis come from a higher response rate observed in Japanese patients compared to Caucasian patients (when treated with gefitinib) two populations with a difference in the frequencies of the EGFR dinucleotide repeat variants. However, given the abundant EGFR expression in skin tissue, and the observed association between skin toxicity and tumor response; the use of surrogate tissue in this instance might be justified. Nonetheless, this issue highlights an important problem in conducting translational work in this field, since obtaining tumor biopsies in prospective trials for hypotheses generation is not a trivial matter for obvious ethical and practical concerns.
However, robust predictive markers are needed in order to identify the relatively small subsets of patients whose tumours are likely to respond to EGFR-targeted therapies. Candidate markers include phosphorylated EGFR, and phosphorylated effector molecules downstream of the EGFR, such as the mitogen-activated protein kinase (MAPK) and protein kinase B (AKT). However, there are concerns about the stability of phosphorylated proteins in primary tumour samples prior to fixation, and protocols for the collection and processing of clinical material for phosphorylated protein analysis have yet to be validated and standardized. More recently, a work shown that KRAS mutation is associated with resistance to cetuximab and a shorter survival in EGFR-positive metastatic colorectal cancer patients treated with this therapy[103]. KRAS mutation status might allow the identification of patients who are likely to benefit from cetuximab and avoid a costly and potentially toxic administration of this treatment in nonresponder patients. Prospective randomized study is needed to validate these results that bring a new possibility of targeted therapy adapted to each patient according to its KRAS mutation status.
Future issues in the development of EGFR inhibitors include the identification of biologic predictors of response, combination with other targeted agents, and their use in earlier stage malignancies.
VEGF as target for anti-angiogenic therapy
The VEGF family comprises six molecules, the best characterized of which is VEGF-A, which is expressed in at least four isoforms derived by alternative splicing. It is a multifunctional cytokine that acts with receptors expressed on the vascular endothelium to render microvessels hyperpermeable to plasma proteins, alters gene expression, induces endothelial cell migration and proliferation and enhances endothelial cell survival, eventually leading to angiogenesis, permeability and protection against endothelial cell apoptosis and senescence[104,105]. VEGFs mediate their functions by binding to one or more of three tyrosine kinase receptors expressed on endothelial cells: VEGF receptor VEGFR-1 (Flt-1), VEGFR-2 (Flk-1 or KDR) and VEGFR-3 (Flt-4). These receptors have tyrosine kinase activity that initiates intracellular signaling on ligand binding[106]. Other receptors identified (neuropilin-1 and -2) are expressed on numerous cell types, but they do not transmit intracellular signals by themselves after ligand binding[107].
VEGF is a major target for antiangiogenic therapy since its overexpression has been associated with vascularity, endothelial cell migration and invasion, poor prognosis and aggressiveness in most malignancies, including CRC[108]. In CRC, the overexpression of VEGF and its receptor correlated with the development of metastasis[109]. Anti-VEGF strategies include neutralizing antibodies to VEGF or its receptors, ribozymes to receptors and TKIs that block downstream signaling despite ligand binding to VEGFR. Several of these strategies are currently under investigation, including PhaseI, II and III trials.
Bevacizumab is a humanized monoclonal antibody that targets and binds to vascular endothelial growth factor-A (VEGF-A), reducing the availability of VEGF and thereby preventing receptor activation[110]. Kabbinavar et al[5] reported the first clinical trial of bevacizumab in combination with 5-fluorouracil and leucovorin (5-FU/LV) in previously untreated colorectal cancer patients. Then, different clinical trials shown that Bevacizumab increases survival in association with chemotherapy in the treatment of metastasic CRC. These data led to the FDA approval of bevacizumab for the treatment of metastatic colorectal cancer in February 2004.
As cetuximab, the development of bevacizumab has not included a diagnostic eligibility test and the identification of biomarkers that may predict which patients are most likely to respond to targeted-therapies is of considerable interest. To date, neither direct measurement of VEGF expression nor assessment of tumor microvessel density has been incorporated into the clinical trials or linked to the rates of response to this antibody.
Possible biologic surrogates which have been tested in some clinical trials include: DCE-MRI, positron emission tomography scan assessment of tumor blood flow[111], mutations in k-ras, b-raf and p53 genes[112], circulating endothelial progenitors, mature circulating endothelial cells[113], or plasma levels of angiogenic markers, e.g.,VEGF, bFGF. To date, few studies have assessed the potential utility of biomarkers in predicting which patients are more likely to respond to antiangiogenic therapy in the clinic. Tumors may express multiple pro-angiogenic factors and, thus, have different pathways to bypass the VEGF inhibition. Likely, biomarkers that summarize the effects of all angiogenic regulators may better predict patient outcome than the analysis of a single angiogenic factor.
FUTURE PERSPECTIVES
Over recent years, a large number of studies have attempted to define molecular and biochemical markers that may be useful predictors of response to treatment. The introduction of DNA microarray technology has revolutionized our approach to understanding the molecular events regulating the drug-resistant, allowing the simultaneous assessment of thousands of genes. This approach provides a valuable means to identify novel biomarkers of response to treatment as well as novel molecular targets for therapeutic intervention. The candidate gene approach has been widely used to identify the genetic basis for pharmacogenetic traits and becomes increasingly more powerful with the recent advances in genomic technologies. The simultaneous testing of multiple markers predictive of response could help to identify more accurately the true role of these polymorphisms in CRC therapy (Figure 5). High-throughput sequencing and SNP genotyping technologies allow the study of thousands of candidate genes and the identification of those involved in drug efficacy and toxicity. Combination of predictive gene sets identified by gene expression profiling with proteomics and SNPs- array methodologies may enhance the prediction of tumor response to chemotherapy and provide further insights into the molecular characterization of tumor cells. In future studies it will important to combine all these technologies to identify the tumoral response to chemotherapy and finally realize an individualized treatment regimen to each patient.
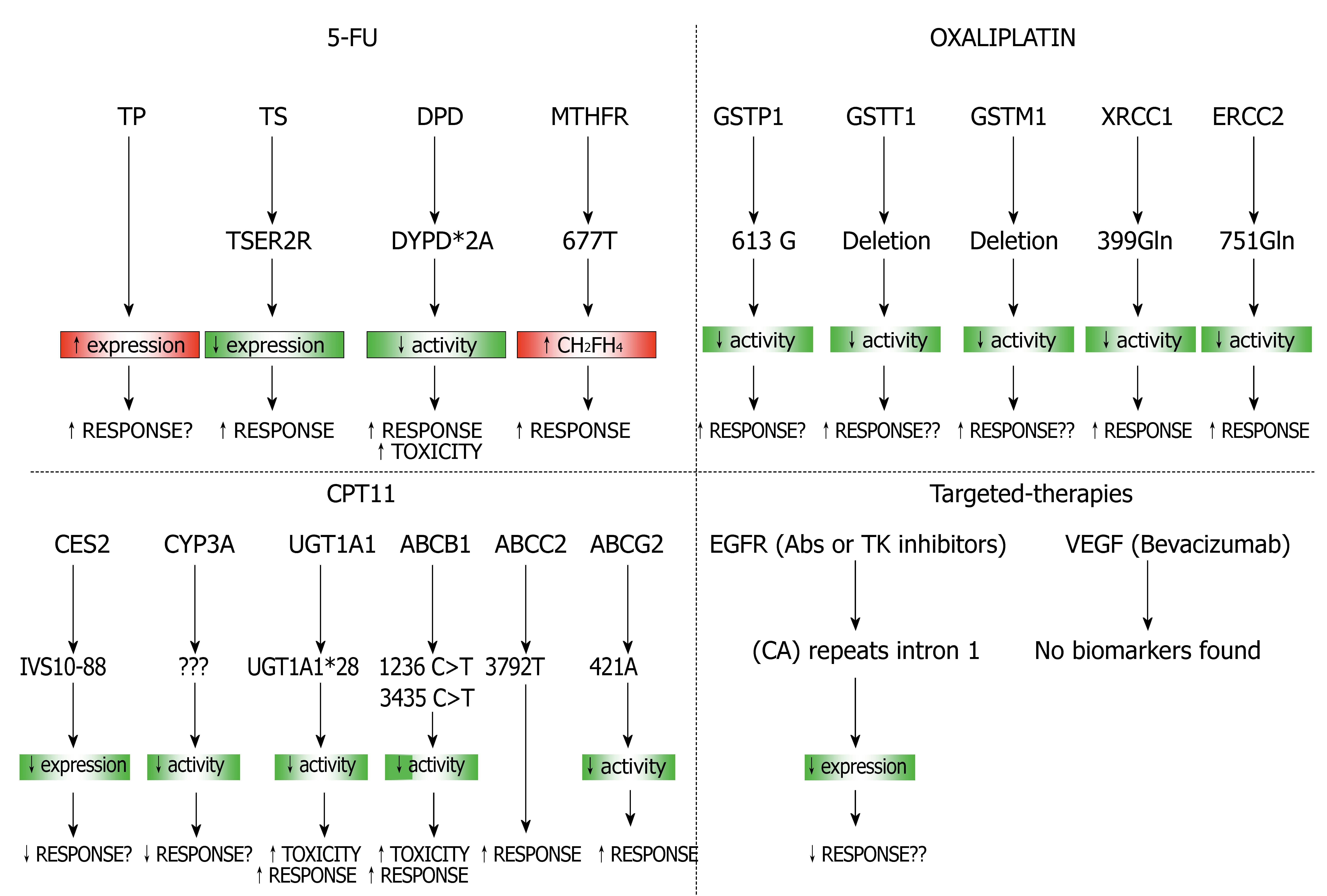
Figure 5 Combination of predictive gene sets for different therapies used in CRC.