MATERIALS AND METHODS
Preparation of plasmid DNA
The HBV replication-competent plasmid used in this study was pHBV4.1, which contains 1.3 copies of the HBV genome (subtype ayw). This construct has been proven to be able to initiate replication of HBV efficiently after being transfected into human hepatoma HepG2 cell lines[5,6]. The plasmid DNA was prepared by using a plasmid-purifying Kit (QIAGEN TIP-500) according to the manufacturer's instructions
Injection of naked plasmid DNA
Male BALB/C mice at specific-pathogen-free (SPF) level, weighing 18-20 g, were provided by the Huaxi Laboratory Animal Center of Sichuan University. Mice were injected via the tail vein with plasmid DNA at different doses in 1.8 mL saline within 5-8 s (hydrodynamic in vivo transfection)[7]. The animals were sacrificed at different time points after injection, and the serum and liver were collected. The serum was stored at -20°C. The liver tissue was separated into two parts: one was fixed in 40 g/L neutral-buffered formalin, and the other was snap frozen in liquid nitrogen and stored at -70°C for subsequent analysis of HBV DNA replication intermediates.
Polyinosinic-polytidylin acid (polyIC) treatment
Twenty-four hours after hydrodynamic injection with pHBV4.1, the HBV replication-competent mice were matched by body weight, age and serum levels of hepatitis B e antigen (HBeAg), and were divided into two groups of five mice each. The mice in the experimental group were injected intraperitoneally with 200 μg polyIC (P-0913; Sigma) in 200 μL phosphate-buffered saline (PBS), three times at 24-h intervals, while the control group mice received only 200 μL PBS. The mice were sacrificed 4-6 h after the final injection of polyIC. Liver tissue was frozen in liquid nitrogen and stored at -70°C for DNA extraction and analysis.
Detection of HBV DNA replication intermediates
Frozen liver tissue was mechanically pulverized in liquid nitrogen and HBV DNA replicative intermediates were isolated as described previously, with some modi-fications[8]. One hundred and twenty micrograms of liver tissue powder was lysed in 0.6 mL 100 mmol/L Tris hydrochloride (pH 8.0) and 2 mL/L NP40. The lysate was centrifuged for 1 min at 14000 rpm in a microcentrifuge. The supernatant was adjusted to 6.75 mmol/L magnesium acetate plus 200 μg DNaseI/mL, and incubated for 1 h at 37°C. The supernatant was readjusted to 10 mmol/L ethylenediaminetetraacetic acid EDTA, 8 g/L sodium lauryl sulfate, 100 mmol/L NaCl and 1.6 mg pronase/mL, and was incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol/chloroform, precipitated with 0.7 volume of isopropanol, and resuspended in 30 μL 10 mmol/L Tris hydrochloride (pH 8.0) and 1 mmol/L EDTA. DNA Southern blot hybridization analysis was performed with 30 μL viral replication intermediates, as previously described[7]. HBV DNA probe was labeled by using DIG Luminescent Detection Kit (Roche Applied Science). A digoxigenin-labeled, full-length HBV DNA probe was used for Southern filter hybridization analysis.
Detection of liver HBsAg and HBcAg by immunohistochemistry
The number and location of cells in the liver that expressed HBV core antigen (HBcAg) or HBsAg were assessed by immunohistochemical staining. Liver tissue fixed in formalin was embedded with paraffin and sectioned (3 μm thick). The paraffin-embedded sections in PBS were treated for 15 min at 37°C with 30 g/L hydrogen peroxide, and washed with PBS. After the sections were blocked with normal goat serum for 20 min at 37°C, rabbit anti-HbsAg (Biodesign) or rabbit anti-HBc (NEOMARKERS) primary antibody was applied at 1:100 or 1:150 dilution for 60 min at 37°C, or overnight at 4°C, respectively. After washing with PBS, the secondary antiserum consisting of Polymer-HRP Anti-Rabbit (Zhongshan, Beijing) was applied at 1:350 dilution for 50 min at 37°C, according to the manufacturer's instructions. The antibody-coated sections were washed with PBS, stained with 3', 3'-diaminobenzidine tetrahydrochloride (DAB), and counterstained with hematoxylin, before being mounted.
Serological analysis for HBV antigen
HBeAg and HBsAg analysis was performed with 50 μL mouse serum by using enzyme-linked immunosorbent assay (ELISA). The HBeAg and HBsAg Detection Kits (Shanghai Shiye Kehua Company, China) were used respectively.
RESULTS
Assessment of the appropriate amount of pHBV4.1 plasmid DNA for hydrodynamic injection
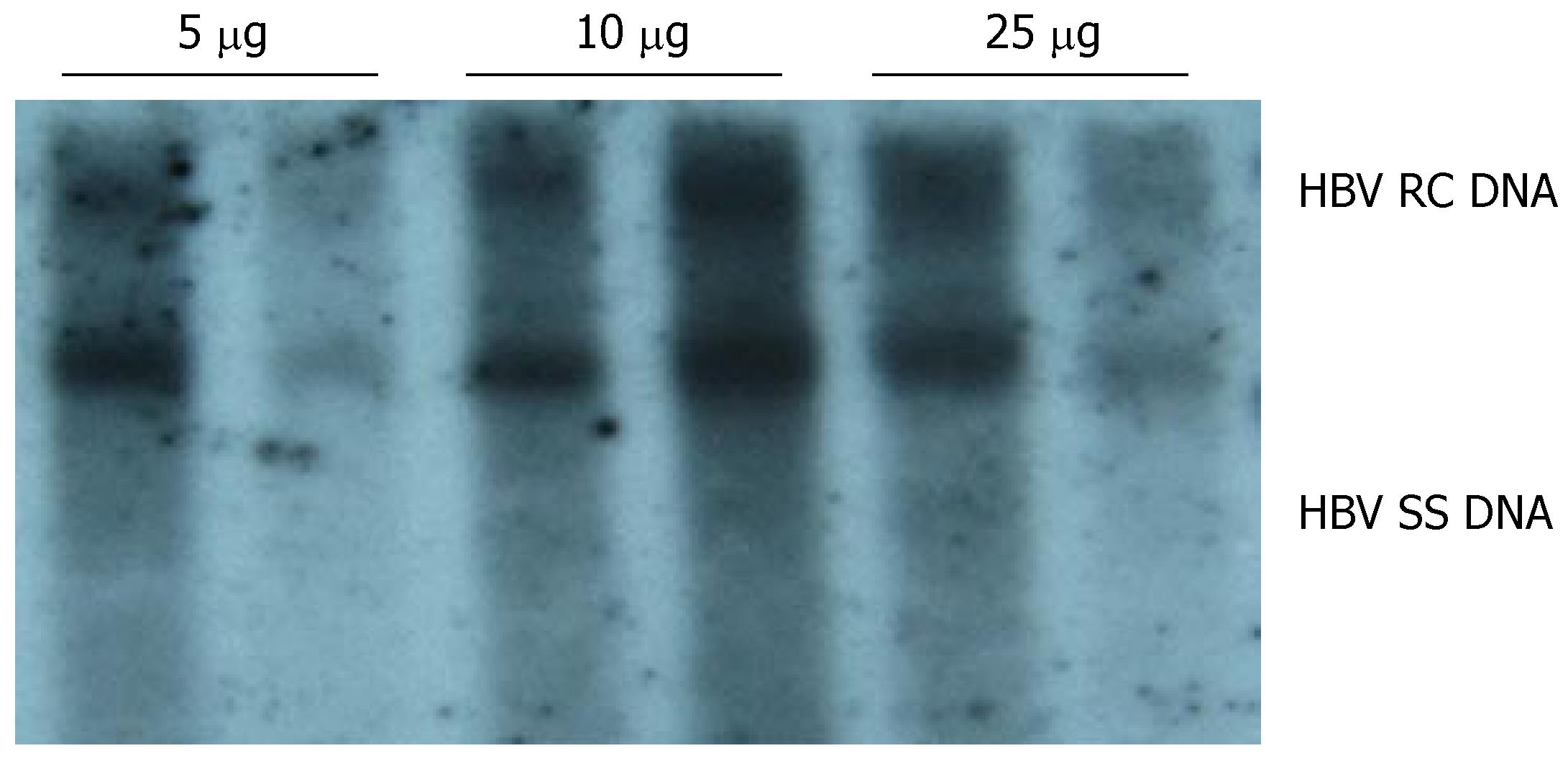
To determine an appropriate amount of pHBV4.1 DNA for establishing the mouse model for HBV replication, mice were divided into different groups (two in each group) and injected with various amount of plasmid DNA (5, 10 and 25 μg per mouse). HBV DNA replicative intermediates were examined on d 4 after injection. The results of three independent experiments showed that a significant level of HBV DNA replicative intermediates was detected in mouse liver when the amount of plasmid DNA injected was as low as 5 μg per mouse. The level of HBV replicative intermediates increased when the amount of plasmid DNA was increased to 10 μg per mouse. A further increase in the amount of plasmid DNA to 25 μg per mouse did not result in a significant increase in HBV replicative intermediates in mouse liver. Therefore, we used 10 μg per mouse as the appropriate dose of pHBV4.1 to develop an HBV-replication-competent mouse model (Figure 1).
Figure 1 DNA-dose-dependent HBV replication.
Various amount of pHBV4.1 in 1.8 mL saline were injected into BALB/c mice within 5-8 s. Mice were sacrificed on d 4 after injection, and HBV DNA intermediates were detected by Southern blotting.
Establishment of a mouse model with HBV replication
Thirty mice were injected with pHBV4.1 plasmid DNA at 10 μg per mouse using a hydrodynamic procedure. These mice were sacrificed on d 1, 3, 4, 5, 7 and 10 after injection (n = 5 at each time point). The level of HBV replication and expression were analyzed.
HBV replication intermediates
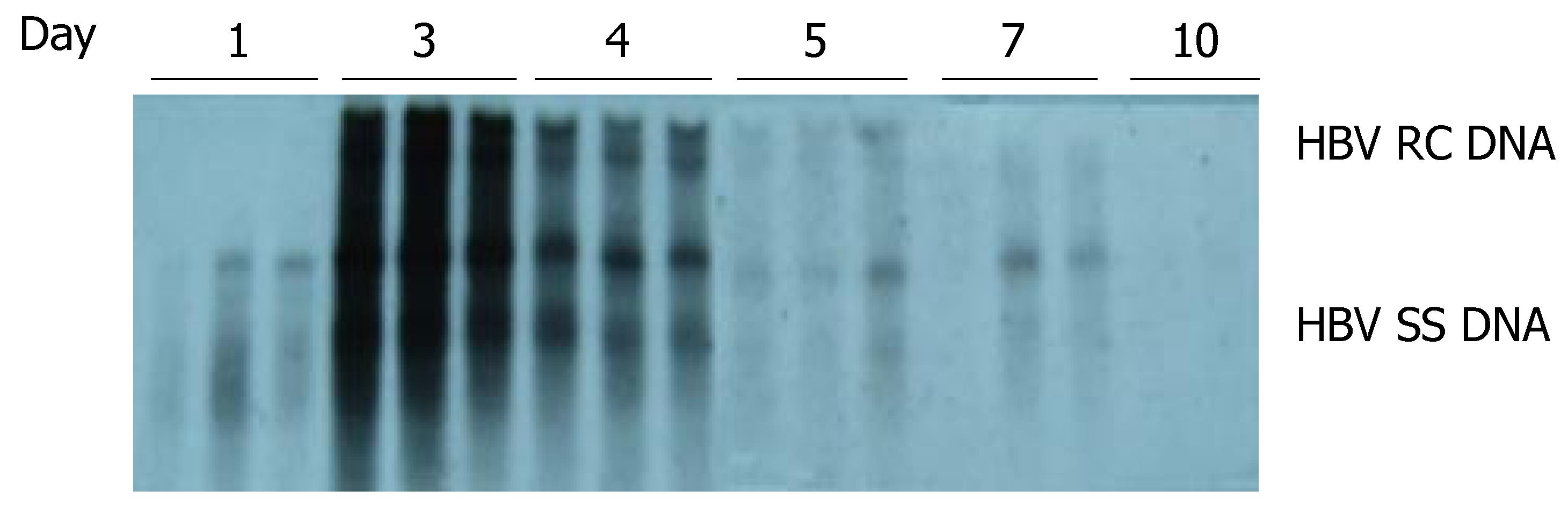
As shown in Figure 2, HBV DNA replication intermediates were detected on d 1 [but without the relaxed circular DNA (RC DNA) form], and were abundant on d 3 and 4. By d 5, the number of HBV replication intermediates decreased, and were eventually undetectable by d 10. These results suggested that HBV could replicate in the liver of the mice, and that the level of HBV replication reached a peak on d 3 and 4 after hydrodynamic transfection.
Figure 2 Time-dependent viral replication after hydrodynamic in vivo transfection with pHBV4.
1. Mice were injected hydrodynamically with 10 μg pHBV4.1 and were sacrificed at different time points after injection. HBV DNA intermediates were detected by Southern blotting.
Expression of HBcAg and HBsAg in mouse liver
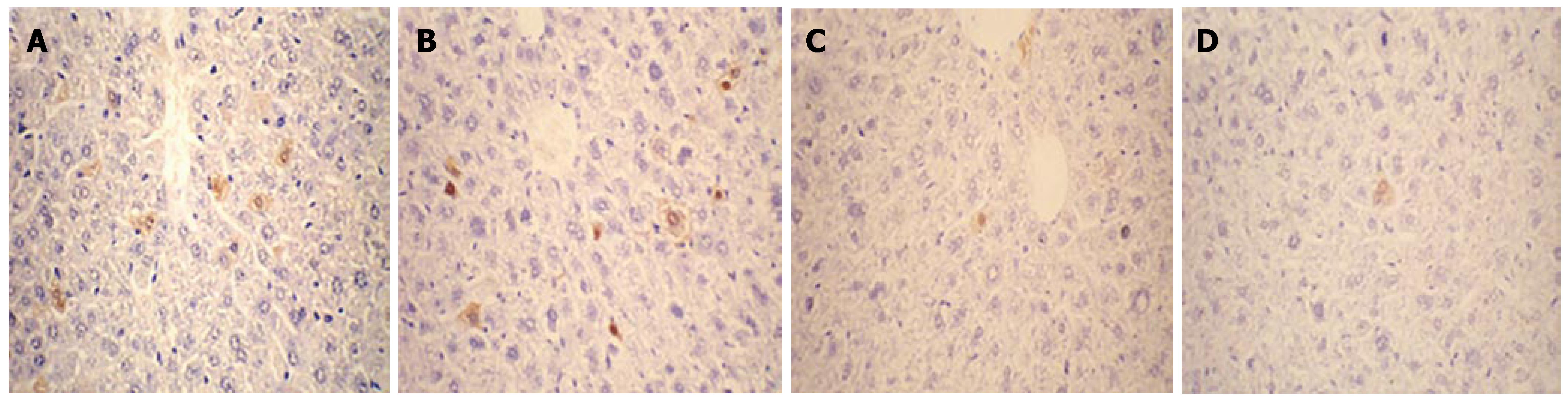
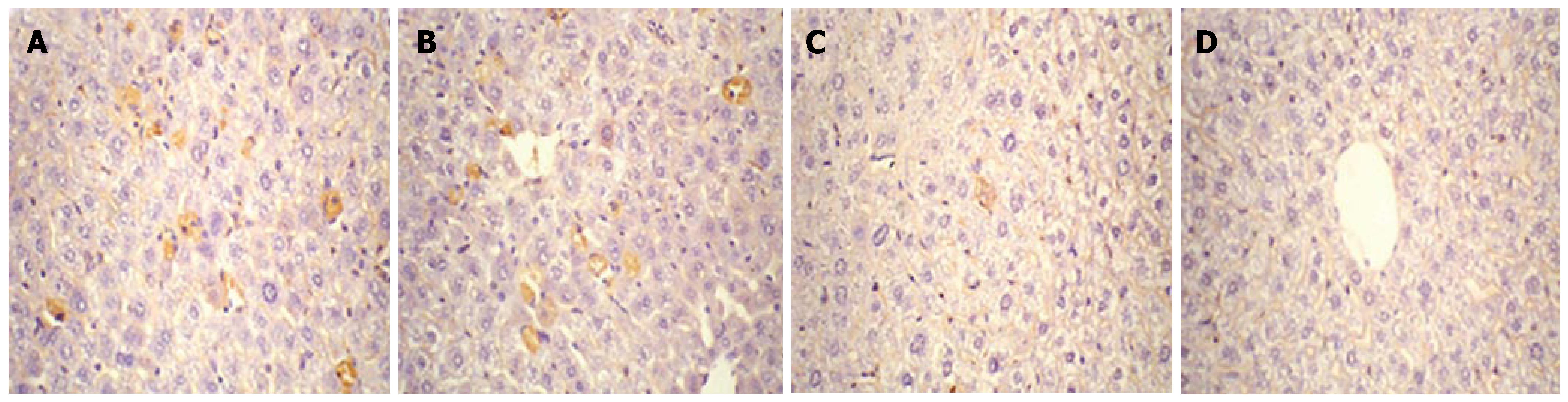
Expression of HBcAg and HBsAg in the liver collected on d 1, 4, 7 and 10 after injection was detected by immuno-histochemistry. As shown in Figures 3 and 4, HBcAg- and HBsAg-positive hepatocytes were randomly distributed throughout the liver lobules, with a tendency for localization in the centrilobular area. HBcAg was present in the nuclei and cytoplasm of the hepatocytes, with most expression in the nuclei. HBsAg staining was only seen in the cytoplasm. The expression of both antigens exhibited a similar time-dependent expression pattern that reached a maximum level on d 1, and remained steady until d 4 after injection, followed by a sharp decline. There were only a few HBcAg- and HBsAg-positive cells in whole liver sections by d 7 and 10.
Figure 3 Detection of HBcAg expression in the liver by immunohistochemical staining (DAB, × 40).
Mice were injected hydrodynamically with 10 μg pHBV4.1 and were sacrificed at different time points after injection and HBcAg in mouse liver was detected. A: 1 d after transfection; B: 4 d after transfection; C: 7 d after transfection; D: 10 d after transfection.
Figure 4 Detection of HBsAg expression in liver by immunohistochemical staining (DAB, × 40).
Mice were injected hydrodynamically with 10 μg pHBV4.1 and were sacrificed at different time points after injection and HBsAg in mouse liver was detected. A: 1 d after transfection; B: 4 d after transfection; C: 7 d after transfection; D: 10 d after transfection.
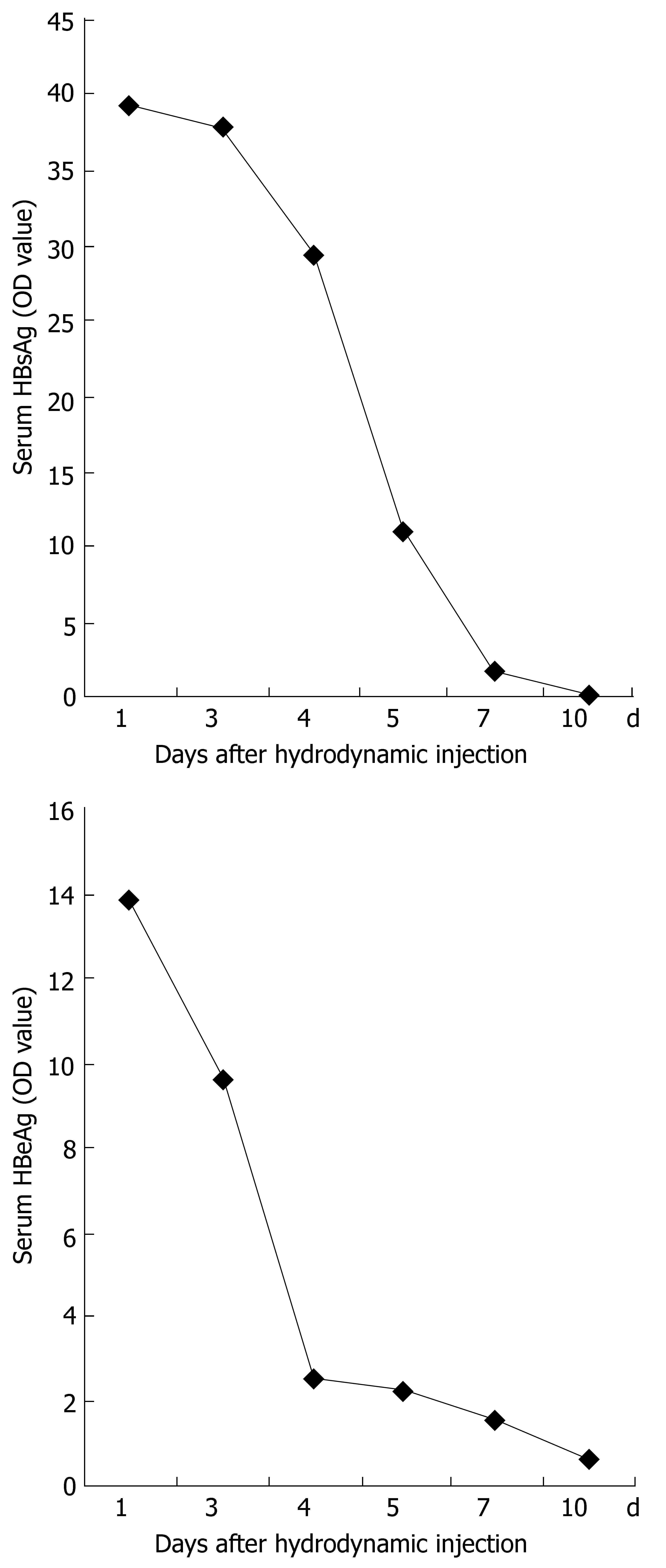
Serum from mice sacrificed on d 1, 3, 4, 5, 7 and 10 after injection was measured by ELISA for the level of HBeAg and HBsAg (OD450). Samples were considered positive at OD450≥ 2.1. The mean level of HBeAg or HBsAg expression at each point was derived from five mice. Figure 5 shows that the expression of HBeAg and HBsAg in the serum was detected on d 1, 3, 4 and 5, reached a peak on d 1 after injection, and decreased thereafter. All samples from mice on d 7 and 10 after injection were negative for HBeAg and HBsAg.
Figure 5 Time-dependent HBV antigen expression in serum after hydrodynamic transfection.
Mice were injected with 10 μg pHBV4.1. At different time points, HBsAg and HBeAg in the serum were measured by ELISA.
HBV replication levels in different parts of mouse liver
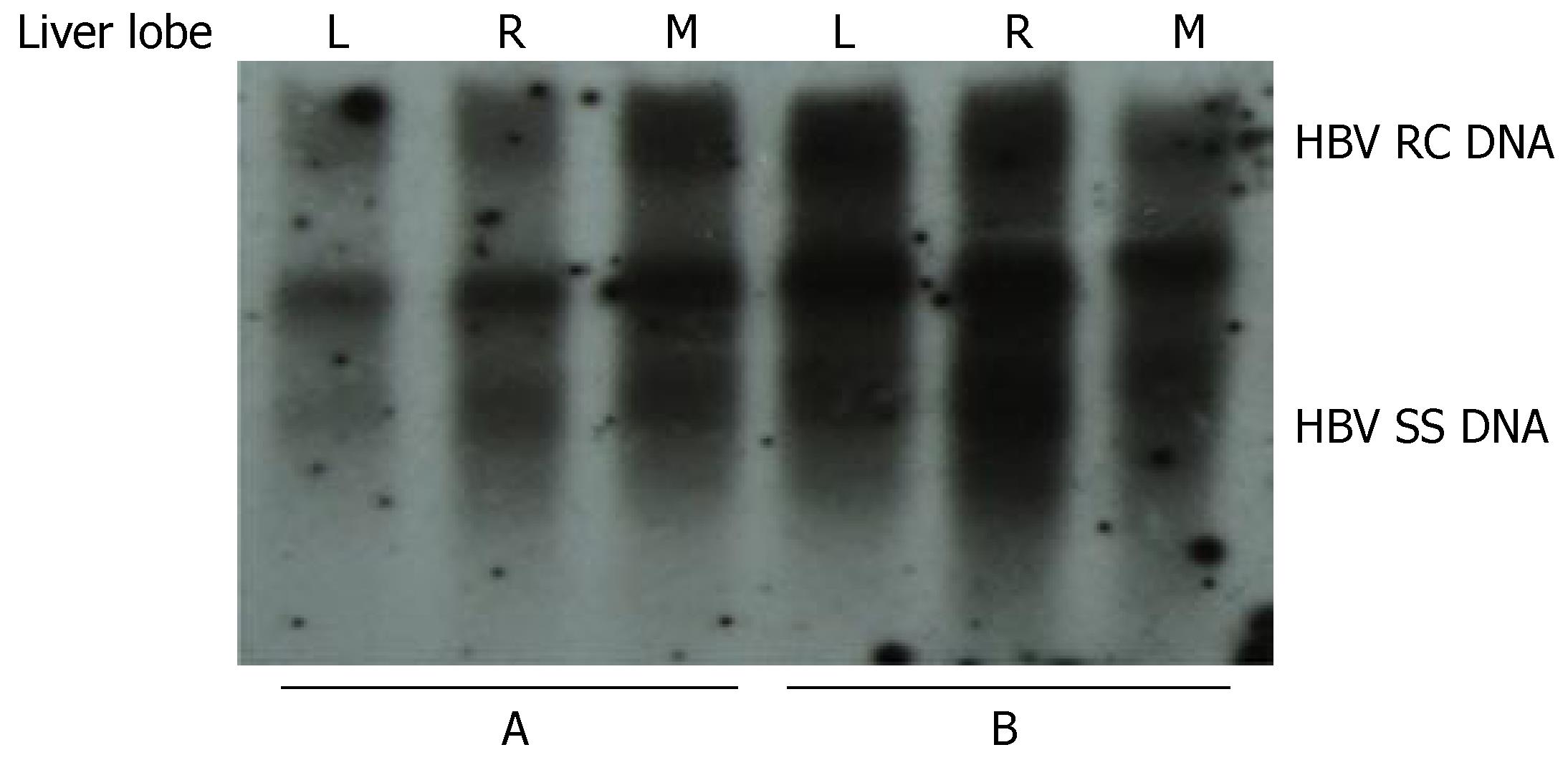
To investigate whether the levels of HBV replication in different parts of the liver are similar, mice were injected with 10 μg pHBV4.1 plasmid DNA and sacrificed 3 d later. The liver of each mouse was divided into three parts: left, right and middle lobules. The level of HBV DNA replication intermediates in each part was analyzed and compared. We found that the level of HBV replication in the left, right and middle lobules in each mouse was similar (Figure 6).
Figure 6 Viral replication in different parts of the mouse liver after hydrodynamic transfection.
The results from two mice (A and B) are shown. L: left lobe; R: right lobe; M, middle lobe.
Effect of polyIC injection on HBV replication
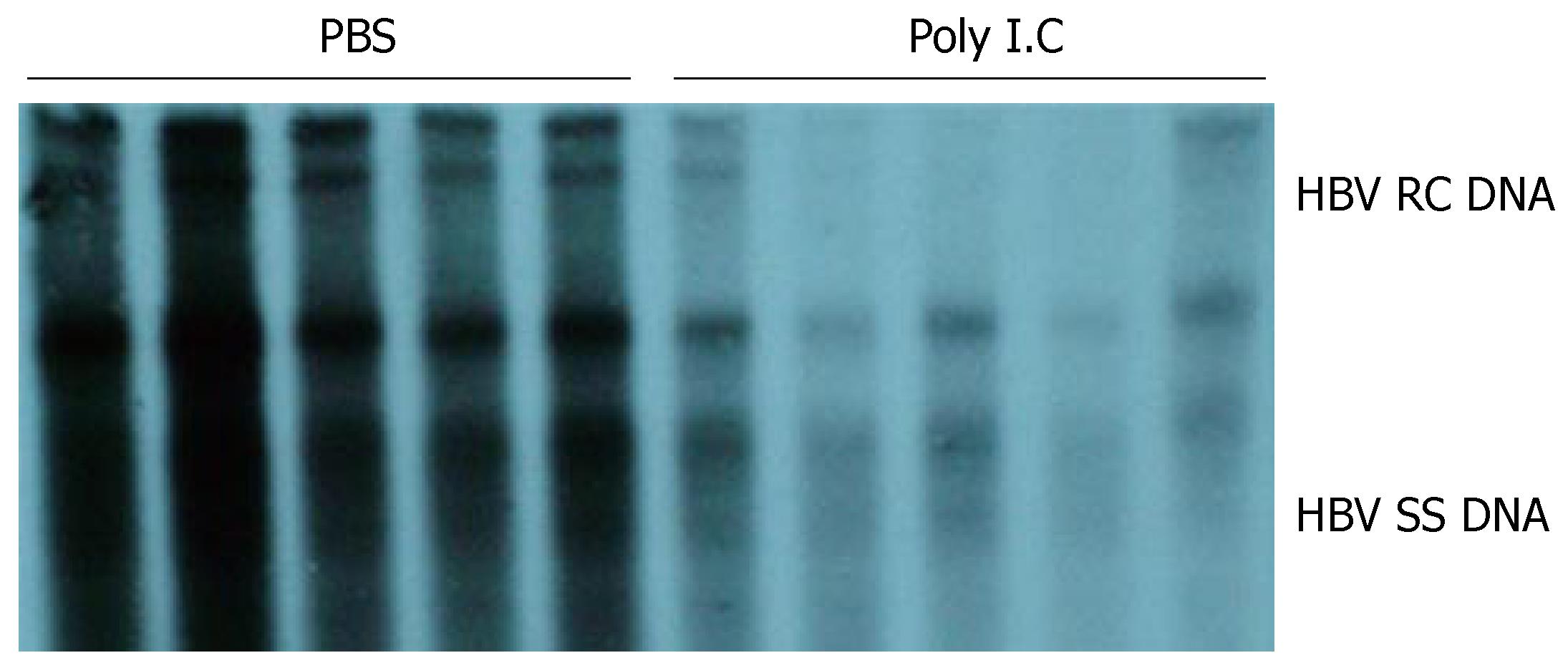
To investigate whether the mouse model established in this study can be used for the study of HBV, we used it to evaluate the effect of polyIC on HBV replication. As shown in Figure 7, compared with the control animals that were injected with PBS only, HBV DNA replicative intermediates dramatically decreased in the liver from mice injected with polyIC. This result suggested that repetitive polyIC injection decreased the level of viral replication in the liver.
Figure 7 Effect of polyIC on HBV replication in mice.
After matched by body weight, age and serum HBeAg, mice were treated with PBS or polyIC. HBV replication levels in the liver of the two groups of mice were compared.
DISCUSSION
HBV is a member of the hepadnavirus family, which contains a group of closely related hepatotropic DNA viruses that infect their respective animal hosts, such as HBV, woodchuck hepatitis virus (WHV)[9] and duck hepatitis B virus (DHBV)[10]. HBV naturally infects human and certain non-human primates such as chimpanzees. However, experiments with chimpanzees are limited by cost, availability and ethical considerations. Also, the ability of HBV to infect tupia and the significance of tupia as an HBV animal model remain controversial. Therefore, much current understanding about HBV is derived from studies of WHV and DHBV infection in their natural host[11]. In addition to their use for the selection of anti-HBV drugs[12,13], they are also used to study the mechanism of HBV replication by reverse transcription[14] and pathogenesis of HBV-related hepatocellular carcinoma[15-17]. However, these two hepadnaviruses have some differences to HBV in terms of genome structure and transcriptional and replicative regulation; therefore, they cannot serve as a substitute for research into HBV[18]. Recently, much attention has been paid to HBV transgenic mice, especially HBV-replication-competent transgenic mice with 1.3 copies of the genome of HBV. It is one of the most widely-used animal models in studies on the mechanism of HBV replication[19-21], the pathogenesis of HBV-related liver diseases[22-24], and the selection of anti-HBV drugs[25-27]. However, the production of transgenic mice is very laborious and time-consuming. Therefore, it is necessary to develop a convenient, rapid and useful animal model for HBV-related studies. Hydrodynamic in vivo transfection is an efficient, rapid and convenient gene-delivery method, invented in 1999[7]. We have reported previously that with the hydrodynamic injection of plasmid DNA encoding HBsAg via the tail vein into mice, efficient expression of HBsAg was detected mainly in the mouse liver, which persisted for about 10 d. In this study, by taking advantage of this hydrodynamic in vivo transfection method, an HBV-replication mouse model was developed by using the HBV-replication-competent plasmid pHBV4.1. The replication of HBV was confirmed by the presence of HBV-replication-intermediate DNA, HBcAg and HBsAg in the mouse liver, and the sera of mice were positive for HBeAg and HBsAg.
For different plasmid DNA, the doses needed for hydrodynamic injection are presumably different; therefore, we first determined the appropriate amount of pHBV4.1 for hydrodynamic transfection. The results of three independent experiments demonstrated that the level of HBV replication was significant and reached a steady-state level when the amount of plasmid DNA injected was as low as 10 μg per mouse. To keep the replication at a high level and also save the plasmid, we used 10 μg per mouse as the appropriate amount of pHBV4.1 to establish an HBV-replication-competent mouse model.
HBV is an enveloped virus containing a 3.2-kb partially double-stranded DNA genome within its nucleocapsids[28,29]. Upon HBV infection of the hepatocyte, the viral genome is converted to covalently closed circular DNA (cccDNA) in the nucleus. The cccDNA serves as the template for transcription by the host RNA polymerase II, which generates the 3.5-, 2.4-, 2.1- and 0.7-kb viral transcriptions that encode the HBV core (HBcAg and HBeAg) and polymerase polypeptide, the large surface antigen polypeptide, the middle and major surface antigen polypeptides (HBsAg)[30-32] and the HBx polypeptide[33]. Besides encoding for the HBV core polypeptide and HBV DNA polymerase that compose the viral capsid, the greater-than-genome-length 3.5-kb pregenomic RNA (pgRNA ) is encapsulated and serves as the template for reverse transcription to synthesize HBV replication intermediates[34-36]. The time-course of the results of our experiments provided the characteristics of HBV replication in the mouse model: within 24 h after transfection, the input DNA apparently reached the nuclei of the hepatocytes, in which it was transcribed and translated to produce HBV-related antigens, and these antigens were soon secreted into the serum. The time at which HBV antigen expression peaked was on d 1 after transfection. The expression level of viral antigen in mouse liver was stable, but the titer of HBeAg and HBsAg in the serum decreased from d 1 to 4 accordingly. The reason for this difference is unclear at present. Viral replication was not robust on d 1, although the presence of small amounts of single-stranded DNA, the earliest replicative DNA intermediates, indicated that viral replication had begun. By d 3 and 4, viral replication reached its peak level and decreased substantially until d 10, which is the time that HBV replication intermediates could not be detected. All these findings suggest that it probably requires about 2 d for 3.5-kb mRNA to produce HBV replication intermediates by reverse transcription abundantly.
The basic mechanism of the hydrodynamic transfection procedure is that when a large volume of DNA solution is injected rapidly via the tail vein, a high hydrostatic pressure develops in the inferior vena cava and causes heart failure, which forces a rapid flow of DNA solution to the liver tissue and DNA molecules into liver cells[7,37-39]. In this way, digestion of the input DNA by DNAase in the blood is greatly reduced.
To assess whether the levels of gene expression in different liver lobes are different, we compared the level of HBV replication in left, right and middle liver lobes from the same mouse. The results suggested that, for a given mouse, the level of HBV-DNA-replicative intermediates was similar in all parts of the liver. This also suggests that the experimental results were not influenced by the use of different liver tissue parts. As shown in Figures 1 and 2, however, there was some variation in HBV replication between different mice, even under similar experimental conditions. This suggests that if this animal model is to be used for selecting anti-HBV drugs or for other research in which HBV replication levels need to be compared, it is necessary to match the HBV-DNA-replication levels by checking serum HBeAg before treatment, and that repeated experiments are needed to guarantee the reliability and accuracy of the test findings.
As a strong inducer of alpha/beta interferon, polyIC has been widely used in HBV transgenic mice and exerts an inhibitory effect on HBV replication[40-42]. Therefore, we applied polyIC treatment to the HBV replication mouse model established in this study to investigate whether this model can be used for studies of HBV. We found that polyIC could inhibit HBV replication, which suggests that the mouse model developed in our study can be used for future research on HBV.
To understand the mechanisms of HBV transcription, replication and pathogenesis, it is usually necessary to mutate the HBV genome, and then measure the transcription, replication and expression levels of viral mutants; in this way, the function of various HBV genome sequences can be understood. If HBV transgenic mice are used, it is necessary for every mutated HBV genome to be transferred into mouse fertilized eggs, and then transgenic mice can be cultivated, but that is very time-consuming, costly and laborious. In contrast, by using the HBV-replication-competent mouse model developed in this study, no surgical techniques or special equipment are needed, which suggests that this model is very simple, convenient, rapid and economical. As a result, this model affords new opportunities for studying HBV; in particular, it is useful for investigating HBV genomic function, especially its role and mechanism in HBV replication through mutation of the HBV genome.
In conclusion, we developed a rapid and convenient mouse model with a high level of HBV replication, and applied it to detecting the inhibitory effect of polyIC on HBV. This provides a very useful tool for functional studies on HBV replication and selection of anti-HBV drugs.
COMMENTS
Background
Hepatitis B Virus(HBV) infection is a worldwide health problem. Because the pathogenic mechanisms of HBV remain unclear, molecular-biological studies for HBV are required. Untill recently, most in vivo studies about HBV have been performed by using HBV transgenic mice. However, the acquisition of transgenic mice is very time-consuming and laborious, Therefore, it is necessary to develop a convenient , rapid and useful animal model.
Research frontiers
HBV mouse model developed by using the hydrodynamic in vivo transfection method is convenient and useful for HBV-related studies. If the mouse genetic background or plasmid backbone are changed, many different HBV mouse models can be developed for different purposes.By using immunodeficient mice or using AAV as backbone of transferred plasmid, chronic HBV carrier mouse models were established.
Innovations and breakthroughs
In this study, we established a mouse model with HBV.replication with naked HBV plasmid through a hydrodynamic procedure and found that for a certain mouse, the levels of HBVDNA replication are similar in left, right, and middle parts of liver tissue. Non-isotope labeled probe and chemiluminescent detection system was used to detect hepatitis B virus replication levels in vivo by southern hybridization.
Applications
This mouse model can be used for functional study of HBV genome, for studies about HBV replication mechanism and for selection of anti-HBV drugs.
Peer review
This paper describes the establishment of a mouse model with hepatitis B virus replication, with the aim to provide a tool for selection of antiviral drugs to inhibit HBV replication. The work is well developed, and shows promising results.