INTRODUCTION
The steady state level of proteins depends on their rates of synthesis and degradation. The proteasome is a major protein-degrading enzyme, which has a predominant role in intracellular protein catabolism. This enzyme influences various cell functions, as the enzyme not only hydrolyzes obsolete and oxidized proteins, thereby preventing cellular toxicity, but also regulates signaling events by destroying short-lived signal transduction factors and by controlling apoptotic cell death. Another important function of the proteasome is the generation of peptides for MHC class I-restricted antigen presentation. Recently, the proteasome has been shown as a potent regulator of the circadian cycle[1] by its role in degradation of Cryptochrome and Period proteins. Besides proteolysis, the proteasome has non-proteolytic functions, regulating transcription, DNA repair and chromatin remodeling[2].
PROTEASOME STRUCTURE AND FUNCTIONS
In mammalian cells, proteasome is localized mainly in the cytosol and the nucleus[3]. It plays an important role in proteolytic centers within the cells, such as the centrosome, a perinuclear structure. Targeted proteins are transported to the centrosome for degradation, which is carried out by two proteasome structures, the 26S and 20S proteasomes. The 26S proteasome consists of core proteasome (20S proteasome) and two 19S (PA700) regulatory particles[4]. The 20S proteasome is a large complex, which has four stacks containing 14 different gene products and is organized as outer alpha (α) subunits that “shape” 20S proteasome and beta (β) subunits that constitute the 20S catalytic core subunits (β1, β2 and β5, or Y, Z and X). β subunits have various activities, based on the ability to cleave either hydrophobic bonds (chymotrypsin-like activity), basic bonds (trypsin-like activity), or acidic bonds (caspase-like activity). The 20S proteasome exists in two forms: constitutive proteasome and immunoproteasome. The immunoproteasome subunits, LMP2, LMP7 and MECL-1, replace the constitutive subunits, usually under the influence of IFNγ[2]. The immunoproteasome is involved in generation of MHC class I-restricted peptides for antigen presentation. The efficiency of peptide cleavage by the immunoproteasome is higher than by the constitutive proteasome[5]. Mice lacking the LMP7 subunit have reduced antigen presentation[6]. Recently, interferon type 1 (IFNα) has been reported as an effective inducer of immunoproteasome in liver cells[7]. Unpublished results from our laboratory confirmed this finding in hepatoma cell lines stimulated with IFNα.
ATP-DEPENDENT AND INDEPENDENT HYDROLYSIS OF PROTEINS BY PROTEASOME
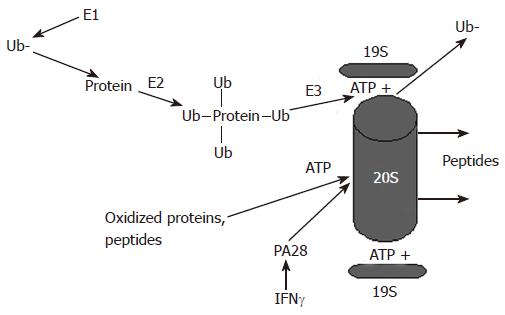
Degradation of proteins by proteasome may or may not require ATP. The 26S proteasome catalyses protein degradation in an ATP-dependent manner, while the 20S proteasome does it ATP-independently. The most important question is how the proteasome “senses” the proteins, which are to be degraded. 26S proteasome usually recognizes proteins, which are marked with ubiquitin, a small 8.5-κDa protein that is covalently attached to protein substrate. To be attached, ubiquitin is activated by E1, the ubiquitin-activating enzyme (Figure 1). Activated ubiquitin is transferred via a high-energy thiol ester intermediate, E2-ubuquitin-conjugating enzyme, to the protein substrate destined for degradation. Protein covalently attaches multiple ubiquitin molecules that generate polyubiquitin chain binding to internal lysine (lysine 48) residue. If the substrate protein binds to monoubiquitin (i.e one ubiquitin molecule per residue) instead of polyubiquitin (K48), it is targeted for degradation by the lysosome. Linkage through Lys 48, is associated with the ubiquitin-proteasome pathway, while Lys 63 linkage has a role in the inflammatory response, endocytic pathway, and ribosomal protein synthesis[8]. The ubiquitin-conjugated protein is subjected for degradation by the 26S proteasome with a help of E3 ligase. To be ubiquitylated and recognized by proteasome, the substrates usually undergo post-translational modification, such as phosphorylation and oxidation. For recognition by E3, in certain cases proteins may require association with molecular chaperones[9].
Figure 1 Proteasome-dependent degradation of proteins by the 26S and the 20S proteasome.
Proteins undergo ubiquitylation and are degraded in an ATP-dependent manner by the 26S proteasome, which activity is regulated by the 19S particle. Alternatively, oxidized proteins can be degraded by the 20S proteasome. This degradation is ATP-independent and is regulated by the 20S proteasome activator, PA28.
The important functions of the 19S particle in 26S proteasome are to recognize ubiquitylated proteins and to open the aperture of the 20S proteasome α-rings to allow substrate entry into the catalytic core. The 19S particle unfolds ubiquitylated substrates, dissociates from ubiquitin with the help of de-ubiquitylating enzymes for further ubiquitin recycling and inserts the proteins into the 20S proteasome for degradation. This insertion needs ATP hydrolysis and is controlled by ATPases, which forms 19S sub-complexes known as the base and the lid.
Alternatively, degradation of non-ubiquitylated proteasome substrates can be carried out in an ATP- and ubiquitin-independent manner by the 20S proteasome, which requires the gate opening by the activators, PA28 α, β and γ (Figure 1). PA28 α and β are predominantly located in the cytosol, whereas PA28 γ has a nuclear localization. PA28 enhances hydrolysis of short peptides. In mice, disruption of PA28 α and β genes leads to defective generation of MHC class-restricted peptides[10], while mice with disrupted PA28 γ gene have slower cell proliferation and enhanced susceptibility to apoptosis. Functions of the PA28 are antagonized by proline-rich proteins, PI31 and Pr39, which, in vitro, block proteasome activity[11,12] 20S proteasome can be simultaneously activated by both 19S and PA28 via the two outer rings. This structure is called a hybrid proteasome, which displays the catalytic properties of both the 26S and 20S forms of the proteasome. These hybrid proteasomes generate a unique spectrum of antigenic peptides for presentation in an ubiquitin-dependent as well as ubiquitin-independent ways[13].
ETHANOL METABOLISM AND LIVER PROTEASOME FUNCTION
The liver is the main site of ethanol metabolism. Hepatocytes express the major ethanol metabolizing enzymes, alcohol dehydrogenase (ADH) and cytochrome P450E1 (CYP2E1), which catalyze the generation of acetaldehyde and reactive oxygen/nitrogen species when liver cells are exposed to ethanol. The level of intracellular oxidative stress depends on the balance between oxidative and protective (antioxidant systems) factors. The level of oxidative stress differentially regulates proteasome function (i.e. low oxidative stress enhances proteasome activity, while high oxidative stress suppresses it).
There is a reciprocal relationship between CYP2E1 and the proteasome. Studies on HepG2 cells that overexpress CYP2E1 showed that ethanol and other CYP2E1 ligands increase the content and activity of CYP2E1 without affecting CYP2E1 mRNA[14,15]. Pulse-chase experiments demonstrated that ethanol and other ligands stabilize the enzyme against proteolysis. The proteasome is responsible for CYP2E1 degradation since treatment with the proteasome inhibitor, PSI, which blocks its chemotrypsin-like activity, caused a dose-dependent increase in the CYP2E1 apoptrotein. Heat shock proteins 70 and 90 provide the communication between CYP2E1 and the proteasome[16,17].
Stabilization of CYP2E1 by ethanol, in turn, results in increased production of ethanol metabolites, which block proteasome function. In vitro treatment of VL-17A cells (ADH/CYP2E1-positive HepG2 cells) with 100 mmol/L ethanol significantly reduces proteasome activity[18,19] as well as the ability of crude proteasome preparations (cytosols) from VL-17A cells to hydrolyze HBV peptide cleavage[20]. The effects of ethanol exposure are blocked by treatment with inhibitors of ethanol metabolism and CYP2E1 activity. In vivo, proteasome inhibition is common in rodents intragastrically fed with ethanol[21,22]. The proteasomal chymotrypsin-like activity reduction in intagastrically fed rats was blunted in animals treated with a CYP2E1 inhibitor, chlormethazole[23]. Furthermore, suppression of proteasome activity by ethanol was established in wild-type mice, but not in ethanol-fed CYP2E1 knockout mice[24]. In orally fed mice, the effect on the proteasome depends on blood ethanol levels achieved after chronic ethanol feeding: usually when blood ethanol exceeds 40 mmol/L, there is a suppression of proteasome function. However, lower blood ethanol levels sometimes result in proteasome activation[21].
The proposed mechanisms for modulation of proteasome activity are related to oxidative modification of proteasomal proteins with primary and secondary products derived from ethanol oxidation. Thus, CYP2E1 activation, reactive oxygen species formation and subsequent adduction of proteasome with protein carbonyls in E47 cells (CYP2E1+ HepG2 cells) caused a decline in the trypsin-like, proteasome activity[25]. Furthermore, in vivo ethanol studies on intragastrically fed rats demonstrated significant induction of CYP2E1 and an increase in 4-hydroxynonenal (4-NHE) concentrations after one month of ethanol feeding, which resulted in decreased proteasome activities[26]. While a 3.5-fold CYP2E1 induction occurred after 15 d of feeding, there was no loss in proteasome activity, indicating that modulation of proteasome activity is a delayed response to CYP2E1-induced proteasome oxidation. Proteasome suppression was attributed to 4-HNE adducts of the 19S (PA700) proteasome regulator specifically an ATPase Rpt4 subunit. It was postulated that this adduction caused disrupted association between the 19S and 20S proteasome particles. In addition, our laboratory discovered the regulating effects of nitration on proteasome function. Nitration is caused by peroxynitrite, a reaction product of superoxide (generated by CYP2E1) and nitric oxide (generated by nitric oxide synthase), which is released as a result of ethanol metabolism and is involved in alcohol-induced liver injury[27]. Massive nitration of 20S proteasome with subsequent 3-NT adduct formation caused a reduction of chymotrypsin-like proteasome activity, primarily due to reduced interaction between PA28 and the 20S proteasome. Interestingly, low levels of nitration activated proteasome function and enhanced proteasome activation by PA28[28]. Similar effects of the peroxynitrite donor, SIN-1, were observed using highly purified proteasome, cultured hepatoma cells and rodent livers. Immunoproteasome has been shown to be more sensitive to the effects of peroxynitrite than the constitutive form of the enzyme[29]. Studies on the 26S proteasome (4-HNE adducts) and on the 20S proteasome (3-NT adducts) revealed that in oxidatively modified proteasome, the ability of 20S proteasome to respond to activating stimuli of its regulators, 19S proteasome and PA28 is limited, which impedes proteasome function. The 26S proteasome is more susceptible to oxidative stress than 20S proteasome[30].
Another potent oxidant generated by ethanol metabolism, acetaldehyde, which is produced by ADH and CYP2E1 has also been shown to affect proteasome function. In vitro adduction of acetaldehyde to cytosolic proteins inhibits proteasome function in a dose-dependent manner[31]. We also observed suppression of proteasome activity in hepatoma cells treated with 400 mmol/L acetaldehyde (Osna, unpublished observation). Thus, not only CYP2E1, but also ADH by generating ethanol metabolites can contribute to proteasome inhibition.
OXIDATIVE MODIFICATION OF PROTEINS: AGGREGATION AND PROTEASOME FUNCTION
The inability for ubiquitylated proteins to be degraded by proteasome is based not only on impaired proteasome activity but also on the relative susceptibility of substrates to degradation. Proteasomal substrates (proteins) can be modified by oxidation of amino acid residues by intracellular oxidants, such as H2O2 or by covalent binding to reactive species, such as peroxynitrite. Depending on their degree of modification, these substrates become either more or less susceptible to proteasome-catalyzed hydrolysis. Thus, when aconitase was treated with peroxynitrite and hydrogen peroxide, low concentrations of oxidants (up to 2 mmol/L for hydrogen peroxide and up to 1 mmol/L for peroxinitrite) enhanced susceptibility of aconitase to degradation, while higher concentrations of oxidants decreased proteolysis[32].
Ubiquitin is essential for ATP-dependent protein degradation by the proteasome. Under certain pathological conditions or due to genetic predisposition, defective ubiquitin + 1 (Ub + 1) is generated by a dinucleotide deletion in Ub mRNA. Proteins ubiquitylated with Ub + 1 are the substrates for proteasomal degradation, but at high concentrations of Ub + 1, the degradation is inhibited[33]. Ub + 1 forms tight noncovalent interactions with target proteins and cannot be removed from the substrate (Luders et al[34], 2003). The accumulation of Ub + 1-labeled undegraded proteins is observed in neurological disorders (e.g. Huntington’s Disease, Alzheimer’s Disease and Down’s Syndrome).
Decreased proteolysis by the proteasome also results in the accumulation of insoluble protein aggregates, which cannot be degraded by proteasome and which further inhibit proteasome function. In patients with alcoholic cirrhosis, the aggregates are known as Mallory bodies or so-called alcoholic hyaline[35,36] and are a common signature of alcoholic liver diseases. Mallory bodies are formed by liver cells when proteasomes are unable to remove cytokeratins, mainly cytokeratins 7 and 19[37]. Experimental studies revealed that the blockade of proteasome with specific inhibitors or by ethanol metabolism induces overexpression of heat shock proteins and Mallory body formation, which was observed both in vivo and in vitro[38,39]. Ub + 1 has also been detected in Mallory bodies, which is a common histological signature of alcoholic liver disease. In vitro delivery of Ub + 1 to mouse hepatocytes caused formation of aggresomes and Mallory body formation[38].
THE UBIQUITIN-PROTEASOME SYSTEM AND ETHANOL-INDUCED CELL DEATH
Proteasome function controls cell death and survival under the normal and pathological conditions. Thus, suppression of proteasome function with specific proteasome inhibitors results in increased cell death. The link between the proteasome function and cell survival is supported by the fact of degradation of certain cell cycle controlling and pro- or anti-apoptotic factors by the 26S proteasome. Because apoptotic events are downstream from proteasome function and ethanol suppresses proteasome activity, it may be possible that proteasome inhibition would promote the accumulation of pro-apoptotic factors in mitochondria of ethanol-metabolizing liver cells. Ultimately, it might stimulate liver cell apoptosis.
Inhibition of the 26S proteasome causes hepatotoxicity in vitro and in vivo. In Hep G2 cells transfected with CYP2E1, the amount of cell death by apoptotic mechanism depended on actual CYP2E1 activity[40]. We also observed elevation of caspase 3 activity in VL-17A cells exposed to 100 mmol/L ethanol[19]. In addition, a proteasome inhibitor, MG132, injected to mice 4 h prior to TNFα injection, sensitized liver cells to TNFα-induced apoptosis[41]. In ethanol non-metabolizing cells, there is a negative feedback between chemotrypsin-like activity of proteasome and caspase-3 activation, when cells were treated with staurosporin. Furthermore, thymocytes, which normally are rich with proteasome, have declined proteasome activity when they underwent apoptosis after dexamethasone treatment[42].
Ethanol promotes cell death by inducing both apoptotic and necrotic events. Mitochondria, the primary site of energy (ATP) generation, are critical for induction of cell death by ethanol. Ethanol metabolism causes mitochondrial dysfunction, including impairment of fatty acid b-oxidation, inhibition of mitochondrial respiration and damage of mitochondrial DNA. The opening of mitochondrial pores (mitochondria permeability transition, MTP), normally blocked by anti-apoptotic factor, Bcl-2, results in release of cytochrome C that initiates the further death events. The scenario of the death events depends on ATP levels. In the case of ATP depletion, cells die via necrosis. However, if ATP levels are not changed, cytochrome C release causes activation of the mediators of apoptosis, caspases, and subsequent DNA fragmentation. Certain pro-apoptotic factors (Bid, Bax) closely associated with the mitochondrial membrane are degraded by the proteasome[43]. Thus, ethanol plays a dual role in regulating cell death: on one hand, it has a tendency to deplete ATP and facilitates necrosis; on the other hand, due to the proteasome suppression, it promotes the accumulation of pro-apoptotic factors in mitochondria of ethanol metabolizing liver cells and initiates apoptosis.
UBIQUITIN-PROTEASOME INVOLVEMENT IN ALCOHOL-INDUCED TISSUE INFLAMMATION
The outcome of acute inflammation depends on the balance of pro- and anti-inflammatory events initiated by mediators of inflammation, which can be “turned on or off”via certain signal transduction pathways. To be effective and non-toxic, the signals must be short enough to allow switching to the next stage of the process (such as the transition from inflammation to regeneration). These changes in gene activation repertoire occur with the help of transduction factors, which, after their job is done, are ubiquitylated and degraded by the proteasome. Alcohol consumption seems to interfere with the transduction of pro-inflammatory signals.
Many important pro-inflammatory and proliferative signals are regulated via the NF-κB pathway. Activation of this pathway occurs by cytokines (such as tumor necrosis factor alpha, TNFα), phosphorylation of cytoplasmic inhibitory protein IkB and subsequent translocation of NF-κB to nucleus to initiate gene transcription. IκB can be “marked” by ubiquitin upon phosphorylation and then is degraded by proteasome; alternatively, non-phosphorylated IκB is modified by small ubiquitin-like modifier (SUMO) and is not targeted for degradation[44]. In ethanol non-metabolizing cells, ethanol enhances NF-κB activation; however, in ethanol metabolizing cells, IκB dissociation is blocked and NF-κB activation does not happen. Acetaldehyde, a product of ethanol metabolism, is responsible for IκB stabilization and non-degradation by the proteasome[45]. Acetaldehyde combines with the product of lipid peroxidation, malondialdehyde, and form the adducts with numerous proteins (malondialdehyde-acetaldehyde adducts, MAA). The MAA adducts reduce the susceptibility of lysozyme to proteasome degradation. MAA-modified proteins can serve as the targets of autoimmune reactions, enhancing liver injury[46].
In liver, ethanol treatment modulates activity of various signal transduction pathways and induces certain downstream events. Thus, feeding of animals for 16 wk with ethanol diet causes up-regulation of chemokines and neutrophil infiltration in the liver. It also induces elevation of IL-8 levels, which results in suppression of the 26S proteasome, due to activated JNK/AP1 pathways[41].
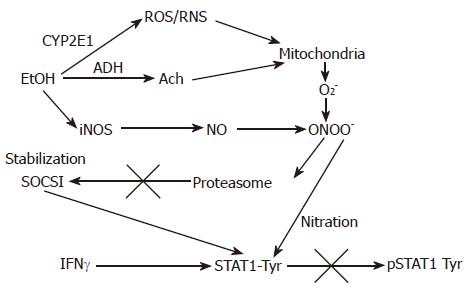
The Jak-STAT pathway, which plays a critical role in inflammation, proliferation, repair, and antiviral defense and usually supports proteasome activation, is also regulated by ethanol metabolism. Thus, acute ethanol exposure inhibited STAT3 activation by leptin in human Huh7 hepatoma cells[47]. Inhibition of MAP kinase, protein kinase A and protein kinase C partially enhance IFN-induced STAT1 phosphorylation reduced by ethanol[48]. We also reported inhibition of STAT1 phosphorylation in VL-17A hepatoma cells[49], which was attributed to ethanol-mediated induction of SOCS1, due to the limited degradation of SOCS1 by suppressed proteasome (Figure 2). Peroxynitrite formation with subsequent nitration of tyrosine residues seems to play a critical role in this process. Similarly, inhibition of proteasome has also been shown to inhibit IL-6 signaling via stabilization of SOCS3[50], which is supposed to be proteasome dependent as well.
Figure 2 The proposed effects of EtOH metabolism on IFNγ-induced STAT1 phosphorylation in VL-17A cells.
EtOH is metabolized by ADH and CYP2E1 to Ach and ROS/RNS and increase a formation of PN (peroxynitrite, ONOO-), a reaction product of O2- and NO, which at high concentrations blocks proteasome function. This causes stabilization of SOCS1, a negative regulator of Jak-STAT1 signaling as well as prevents STAT1 phosphorylation on tyrosine residues.
Except liver tissue, some other types of tissues (brain, heart) induce the inflammation in response to ethanol and its metabolism. In these tissues, ethanol metabolism occurs predominantly via CYP2E1 and catalase, a peroxisomal enzyme, which catalyze ethanol oxidation by using H2O2 as a hydrogen acceptor. Hepatocytes transfected with catalase, are also protected from lipid peroxidation after glutathione depletion[51]. In all ethanol-affected tissues, ethanol can also undergo non-oxidative metabolism and form fatty acid ethyl esters, which also induce mitochondrial dysfunction, inhibit protein synthesis and toxically affect cultured cells[52].
ETHANOL-ELICITED IMPAIRMENT OF PROTEASOME FUNCTION AND MHC CLASS I-RESTRICTED ANTIGEN PRESENTATION
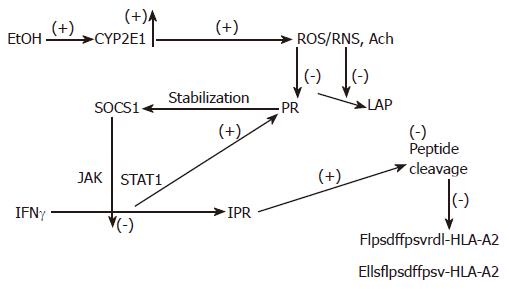
Processing of peptides for MHC classI-restricted antigen presentation is necessary for elimination of liver cells that express viral and other non-self proteins. At the most downstream level, these non-self proteins are cleaved by the proteasome in the cytoplasm[10,53]. Further N-terminal cleavage is carried by aminopeptidases[54]. These cleavages are regulated by IFNγvia the Jak-STAT1 pathway. Generated 8-10 amino acid peptides are presented on the cell surface, for recognition by cytotoxic T-lymphocytes (CTLs) and elimination. Escaping surveillance of infected cells by CTLs ultimately causes chronic persistence of viral infection in the liver, as is often observed in case of viral hepatitis B and C. Recently, we have shown that ethanol treatment of VL-17A cells suppresses proteasome and leucine aminopeptidase functions, which results in a partial blockade of HBV peptide hydrolysis and suppression of IFNγ-induced expression of PA28 and immunoproteasome[20] (Figure 3). Ethanol-elicited impairment in IFNγ signaling is partially responsible for the latter event. This situation might be observed in chronically HBV-infected alcohol consumers, who are unable to clear viral infection in the liver.
Figure 3 The proposed mechanism of regulation of peptide cleavage by ethanol metabolism.
Ethanol (EtOH) treatment induces CYP2E1 activity, which catalyzes production of oxidants. These products block proteasome (Pr) and leucine aminopeptidase (LAP) activities, stabilizing SOCS1 and preventing IFNγ-dependent activation of PA28 and immunoproteasome (IPr). Finally, generation of C-and N-extended peptides for MHC class I-restricted antigen presentation is blocked.
CONCLUSION
Ethanol exposure creates oxidative stress in liver cells and that bi-phasically affects proteasome activity: low oxidative stress up-regulates proteasome function, while high oxidative stress suppresses it. Both acetaldehyde and CYP2E1-induced ROS and RNS contribute to this inhibition. Suppression of proteasome function results in enhanced cell death, mainly due to increased apoptosis. It also induces inflammation, by modulating various signal transduction pathways. Low proteasome activity interferes with efficient MHC class I-restricted presentation of antigenic peptides, which lowers antiviral defense.