Published online Aug 7, 2007. doi: 10.3748/wjg.v13.i29.3990
Revised: October 15, 2006
Accepted: November 4, 2006
Published online: August 7, 2007
AIM: In order to characterize the qualitative and quantitative microorganisms in different sites of the lower digestive tract (LDT) in healthy volunteers, a specific technique was developed for collecting mucous of the distal ileum, colon and rectum.
METHODS: A polyethylene tube was designed to go through the colonoscope channel with a No. 8 French tube. In order to avoid internal contamination, the distal extremity was protected with a membrane of microfilm after being sterilized in ethilene oxid. To facilitate the aspiration of a precise volume, its interior was coated with silicone. One hundred microlliter (0.1 mL) sample of mucous was collected and transferred into an Eppenddorff tube containing nine hundred microlliter (0.9 mL) of VMGA-3 (viable medium of Goteborg). This procedure was repeated at each site of the LDT with a new sterilized catheter.
RESULTS: All sites revealed the “non pathogenic” anaerobic bacteria Veillonella sp (average 105 colony forming units/mL-CFU/mL), allowing to conclude an environment of low oxidation-reduction potential (redox) in the LDT. It was also characterized the presence of Klebisiella sp with significant statistical predominance (SSP) in the ileum. Enterobacter sp was found with SSP in the sigmoid colon, Bacteroides sp non-pigmented (npg) and E.coli with SSP in the sigmoid colon and rectum, Enterococcus sp and Lactobacillus sp with SSP in the rectum, all in a mean concentration of 105 CFU/mL.
CONCLUSION: This procedure is feasible and efficient and can point out a similar distribution of the aerobic and anaerobic bacteria with the presence of biological markers of normal microbiota in the LDT.
- Citation: Quintanilha AG, Zilberstein B, Santos MAA, Pajecki D, Moura EGH, Alves PRA, Maluf-Filho F, Cecconello I. A novel sampling method for the investigation of gut microbiota. World J Gastroenterol 2007; 13(29): 3990-3995
- URL: https://www.wjgnet.com/1007-9327/full/v13/i29/3990.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i29.3990
Indigenous microbiota is highly competitive with other microorganisms in multiplication[1]. Experimental studies have shown the importance of this microbiota (anaerobes only) to avoid colonization of transitory microbiota[2], which is more effective than the protection provided by immunological mechanisms[3]. However, these studies were limited to the analysis of only the anaerobic flora, some segments of the colon utilizing feces samples or luminal content. Additionally, it is difficult to identify and quantify anaerobic and/or yeast microorganisms involved in infectious processes.
The social and practical benefit of investigating indigenous microbiota is to enable investigators to improve prophylactic/therapeutic methods when antimicrobials are used to determine biological markers and to establish a normal pattern utilized for the analysis of microbiological behavior in different diseases of the lower digestive tract.
The aim of this study was to analyze prospectively the indigenous microbiota (qualitative and quantitatively) at different sites of the lower digestive tract in healthy volunteers with a standardized method of collecting intestinal mucous after bowel preparation for colonoscopy for routine utilization in screening patients.
This prospective study was performed with the participation of the Microbiological Laboratory of the Digestive Surgery Division and the Gastrointestinal Endoscopy Unit of the Hospital das Clínicas of the São Paulo University - School of Medicine, and Institute of Biomedical Sciences of the São Paulo University. The protocol was approved by the Ethics Committee of the Institution with the support of the São Paulo’s State Foundation for Research Support (FAPESP).
The study included 24 healthy volunteers (15 females and 9 males) with their age ranging from 18-70 (mean age 53) years.
All the volunteers gave their written informed consent.
Samples were taken only from those whose colonoscopies were normal. The volunteers were not on antibiotics and/or anti-inflammatory medications during the last six months and had no prior abdominal surgery, history of diabetes, scleroderma and/or cancer.
Liquid diet and intake of four Bisacodil pills were recommended at the night before examination. On the day of examination, an adequate volume of 500 mL/L manitol at 20%[4] diluted in 500 mL of orange juice was given until the stools became liquid and clear with no residues. Before examination the patient was sedated with diazepam (up to 10 mg) and meperidine (up to 100 mg).
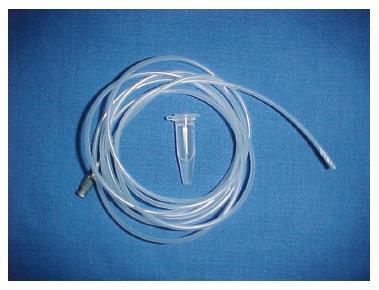
Colonoscopy was performed with an Olympus video-colonoscope using two channels. For collecting mucous, a catheter specially adapted and developed for this study, was utilized (Figure 1).
The catheter was made from a polyethylene No.8 French tube. Silicone (Repel-Silane ES, Pharmacia Biotech) was applied in the lumen of the catheter, to allow and facilitate a continuous column aspiration of one hundred microlliter (0.1 mL) of mucous.
The catheter was constructed with a special protection in the distal extremity, consisting of a microfilm membrane, which was disregarded when the distal extremity reached the lumen of the chosen site. This device was developed in order to avoid contamination of the catheter during its passage through the operation channel of the colonoscope and also to facilitate its liberation at the moment of collecting samples. The catheter’s distal extremity was marked to orient the right volume (0.1 mL of mucous) to be collected with a syringe connected to its proximal end. To collect mucous in each of the predetermined seven sites of the LDT, a new catheter, previously sterilized in ethylene oxide was utilized; therefore seven different catheters were utilized in each volunteer[5,6].
This methodology did not increase significantly the colonoscopy duration with its time being similar to a routine examination.
Colonoscopy was performed at least five hours after colonic preparation. For collecting mucous samples, the catheter was introduced through the colonoscope operating channel. Meticulous care was taken not to contaminate the proximal extremity. After the catheter position was checked, 10 mL of air was introduced with a sterile syringe to disclose the microfilm membrane, which was then liberated. The colonoscope was pulled back slightly and one hundred microlliter (0.1 mL) of mucous on the opposite wall of the viscous was collected. This measurement was oriented by the mark on the distal extremity of the catheter.
The entire catheter was withdrawn and 0.1 mL of mucous was injected into the Eppenddorff tube with its external surface disinfected with alcohol before use. This mucous was gently mixed with 0.9 mL of VMGA-3 (viable medium of Goteborg) solution[7]. Each procedure was repeated seven times, and serial samples were collected from the terminal ileum, cecum, colons (ascending, transverse, descending and sigmoid) and rectum. The samples prepared as described above, were sent to the Microbiological Laboratory, for dilution and plating within one hour at most.
One hundred microlliter (0.1 mL) of mucous samples from each serial dilution (10-1-10-9) was plated and cultured for microaerophylic, aerobic, anaerobic bacteria and yeast utilizing Chapman-Stone medium (DH Co. St. Louis, MO, USA), MacConkey agar (DH Co. St. Louis, MO, USA), Columbia blood agar (Merck Diagnostica, RJ, Brazil), Sabouraud-agar(Merck Diagnostica, RJ, Brazil), selective Enterococcus agar (Merck Diagnostica, RJ, Brazil), phenylethyl alcohol agar (DH Co. St. Louis, MO, USA), Veillonella medium (DH Co. St. Louis, MO, USA), BHI (DH Co. St. Louis, MO, USA) + K vitamin + haemin + streptomycin, reinforced Clostridium medium (Merck Diagnostica, RJ, Brazil), Bacteroides fragilis bile-esculin agar medium (BBE), Bifidobacterium medium, Propionibacterium medium, BHI (DH Co. St. Louis, MO, USA) + yeast extract (2.5 mL/L).
After incubation, the microorganisms (bacteria and/or fungi) were identified and quantified[8].
In order to confirm the distribution of each bacterium in different regions of the LDT, chi square (χ2) test was employed. The expected frequency (EF) of each bacterium in each region of the LDT was calculated using non-parametric tests[9], therefore the data were referred by the median, maximum and minimum values.
The cultured results were expressed as colony forming units/milliliter (CFU/mL) in logarithm base 10 (Log10). P < 0.05 was considered statistically significant.
The distribution of microorganisms in the LDT was identified and quantified (Table 1).
| Microorganisms | Colon | |||||||||||||
| Ileum | Cecum | Ascending | Transverse | Descending | Sigmoid | Rectum | ||||||||
| MC | % | MC | % | MC | % | MC | % | MC | % | MC | % | MC | % | |
| Bacillus sp | 1 | 9.5 | 2 | 4.2 | 5 | 4.2 | 3 | 4.2 | 1 | 4.3 | 3 | 5 | ||
| Bacteroide sp (pig.) | 5 | 4.8 | 5 | 4.3 | 1 | 5 | ||||||||
| Bacteroides sp (npg) | 3 | 47.6 | 5 | 29.2 | 4 | 37.5 | 5 | 12.5 | 5 | 12.5 | 5 | 47.8 | 5 | 65 |
| Bacteroides sp | 2 | 4.8 | 2 | 4.2 | 3 | 5 | ||||||||
| Bifidobacterium sp | 2 | 9.5 | 3 | 4.2 | 4 | 4.2 | 4 | 8.3 | 4 | 8.7 | 3 | 5 | ||
| Candida sp | 3.5 | 9.5 | 2 | 20.8 | 3 | 20.8 | 2 | 29.2 | 4 | 16.7 | 3 | 34.8 | 3 | 35 |
| Clostridium rammosum | 1 | 4.8 | 3 | 4.2 | ||||||||||
| Clostridium sp (gel -) | 4 | 66.7 | 4 | 45.8 | 5 | 50 | 4.5 | 58.3 | 5 | 54.2 | 5 | 69.6 | 7 | 60 |
| Clostridium sp (gel+) | 2 | 4.2 | 4 | 4.2 | 3 | 5 | ||||||||
| Clostridium sp | 1 | 4.8 | 4 | 45.8 | 7 | 4.2 | 5 | 5 | ||||||
| Corynebacterium sp | 3 | 57.1 | 3 | 54.2 | 3.5 | 50 | 4 | 41.7 | 4 | 45.8 | 5 | 60.9 | 5 | 65 |
| E.coli | 5 | 47.6 | 5 | 37.5 | 5 | 45.8 | 4.5 | 50 | 5 | 37.5 | 5 | 69.6 | 7 | 80 |
| Enterobacter cloacae | 4 | 4.2 | 1 | 4.2 | ||||||||||
| Enterobacter sp | 4 | 28.6 | 4 | 20.8 | 7 | 37.5 | 4 | 29.2 | 5 | 45.8 | 5 | 52.2 | 7 | 35 |
| Enterococcus faecalis | 4.5 | 9.5 | 3 | 4.2 | 4 | 4.2 | 3 | 4.2 | 1 | 4.2 | 4 | 8.7 | ||
| Enterococcus faecium | 5 | 4.2 | ||||||||||||
| Enterococcus sp | 4 | 38.1 | 2.5 | 33.3 | 3.5 | 33.3 | 3 | 58.3 | 5 | 58.3 | 5 | 34.8 | 5 | 60 |
| Eubacterium lentum | 4 | 4.8 | ||||||||||||
| Eubacterium sp | 3.5 | 9.5 | ||||||||||||
| Fusobacterium fusiformes | 3 | 4.8 | ||||||||||||
| Fusobacterium sp | 3.5 | 19 | 2.5 | 25 | 3 | 20.8 | 4 | 16.7 | 4 | 8.3 | 3 | 21.7 | 4 | 55 |
| Klebsiella pneumoniae | 5 | 4.2 | ||||||||||||
| Klebsiella sp | 5 | 76.2 | 4 | 54.2 | 6 | 62.5 | 5 | 54.2 | 5 | 41.7 | 7 | 69.6 | 7 | 65 |
| Lactobacillus acidophillus | 4 | 4.8 | 3 | 4.2 | ||||||||||
| Lactobacillus sp | 4 | 33.3 | 2 | 29.2 | 2.5 | 33.3 | 3 | 37.5 | 3 | 25 | 2 | 43.5 | 4 | 70 |
| Leptotrichia sp | 1 | 4.2 | 1 | 4.3 | ||||||||||
| Peptococcus anaerobius | 2 | 4.8 | 4 | 4.2 | 2 | 4.2 | 1 | 4.3 | ||||||
| Peptococcus assachalyticus | 2 | 4.8 | ||||||||||||
| Peptococcus sp | 2 | 28.6 | 2.5 | 33.3 | 3 | 25 | 3 | 33.3 | 3 | 25 | 3 | 56.5 | 3 | 35 |
| Peptostreptococcus sp | 3 | 9.5 | 2 | 8.3 | 1 | 4.2 | 5 | 13 | ||||||
| Propionibacterium sp | 3 | 52.4 | 3 | 20.8 | 4 | 33.3 | 3 | 50 | 3 | 29.2 | 5 | 26.1 | 5 | 30 |
| Proteus | 3.5 | 19 | 4 | 12.5 | 5 | 33.3 | 5 | 37.5 | 5 | 25 | 7 | 34.8 | 7 | 30 |
| Pseudomonas sp | 3 | 9.5 | 1 | 4.2 | ||||||||||
| Rodothorula sp | 4 | 14.3 | 2 | 20.8 | 1 | 4.2 | 1 | 12.5 | 3 | 16.7 | 1 | 4.3 | 3.5 | 20 |
| Selenomonas sp | 2 | 5 | ||||||||||||
| Staphylococcus sp | 5 | 4.8 | 2 | 4.2 | ||||||||||
| Staphylococcus sp (coag -) | 2 | 33.3 | 2 | 20.8 | 1.5 | 8.3 | 2 | 20.8 | 2 | 8.3 | 3 | 21.7 | 3 | 45 |
| Staphylococcus sp (coag +) | 3 | 5 | ||||||||||||
| Streptococcus sp (coag - ) | ||||||||||||||
| Streptococcus (gama hem) | 2 | 8.3 | ||||||||||||
| Streptococcus sp (alfa hemolítico grupo Viridans) | 4 | 14.3 | 4 | 16.7 | 2 | 8.3 | 3.5 | 8.3 | 2 | 13 | 3 | 5 | ||
| Veillonella sp (Gel - ) | 3 | 8.3 | ||||||||||||
| Veillonella sp | 4 | 90.5 | 2.5 | 83.3 | 4 | 70.8 | 4 | 62.5 | 5 | 75 | 5 | 95.7 | 5 | 90 |
In the Ileum 36 genera were identified predominating in a mean concentration (MC) higher than 104 (CFU/mL): 66.7% Clostridum sp, 76.2% Klebisiella sp and 90.5% Veillonella sp. Klebisiella sp was the most prevalent microorganism.
In the cecum 26 genera were identified predominating in a MC higher than 104 (CFU/mL): 54.2% Klebisiella sp and Corynebacterium sp, 45.8% Clostridium sp (gel-) and 83.3% Veillonella sp, with a mean concentration of 102.5 CFU/mL.
In the ascending colon 29 genera were identified predominating in a MC higher than 104 CFU/mL; 50% Clostridium sp (gel-), 62.5% Klebisiella sp, 70.8% Veillonella sp.
In the transverse colon 42 genera were identified predominating in a MC higher than 104 CFU/mL: 58.3% Clostridium sp (gel-), 62.5% Veillonella sp.
In the descending colon 23 genera were identified predominating in a MC higher than 104 CFU/mL: 54.2% Clostridium sp (gel-), 58.3% Enterococcus sp and 75.0% Veillonella sp.
In the sigmoid colon 24 genera were identified pre-dominating in a MC higher than 104 CFU/mL: 69.6% Clostridium sp, E.coli and Klebiella sp, 95.7% Veillonella sp. The Bacteróides sp (npg), E.coli, Enterobacter sp and Candida albicans had a statistically higher prevalence when compared to the other sites.
In the rectum 28 genera were identified predominating in a MC higher than 104 CFU/mL: 70.0% Lactobacillus sp, 80.0% E.coli, 90.0% Veillonella sp. Bacteróides sp (npg), E.coli, Enterococcus sp, Lactobacillus sp and Candida albicans had a statistically higher prevalence when compared to the other sites.
Bacteroides sp (npg) was found at all sites. However, it was more prevalent in the ileum, rectum and sigmoid colon in MC 105 CFU/mL. The Clostridium sp (gel-) was found at all sites in more than 50% of the cases, with its MC higher than 104 CFU/mL, except in the cecum. Corynebacterium sp was found at all sites with varied MC. E.coli was found at all sites and was statistically higher in the sigmoid colon and rectum, with MC higher than 105 CFU/mL. Klebisiella sp and Veillonella sp were found at all sites, with Klebisiella sp being statistically higher in the ileum with MC 105 CFU/mL, and Veillonella sp with MC higher than 104 CFU/mL, except in the cecum with MC of 102.5 UFC/mL.
Lactobacillus sp was found at all sites with a low prevalence and MC, being higher in the rectum. Enterobacter sp, Enterococcus sp and Proteus sp had a low prevalence at all sites, however with high MC when present. Enterobacter sp was statistically higher in the sigmoid colon, with its MC higher than 104 CFU/mL. Enterococcus sp was statistically higher in the rectum, with its MC being 105 CFU/mL.
Candida sp was found with a low MC and prevalence at all sites, being statistically higher in the sigmoid colon and rectum with its MC being 103 CFU/mL. Most of these genera were found in the rectum. Klebisiella sp, Clostridium sp (gel-) and Veilonella sp were detected at all sites with a high MC and prevalence. The main bacterial genera in each segment are shown in Table 2.
The development of reproducible and reliable sampling methods for microbiological studies of the LDT has been a challenge for many years[10,11]. Shinner[12] developed a stainless steel capsule which aspirates the jejunal content, but its use is very complicated. Kalser et al[13] have designed a double lumen polyvinyl catheter with a mercury weight at its distal extremity to obtain samples starting 75 cm from the Treitz ligament to the proximity of the ileo-cecal valve.
Similar methods have been utilized by various authors in the study of bacterial translocation in critical patients. The investigation of Belov et al[14] is outstanding. They evaluated the levels of sepsis mediators (TNF and IL-1) in the jejunal aspirate from patients in septic shock.
The flora study of the LDT represents a greater challenge due to the great concentration and variety of microorganisms in this region. The objective of the first attempts is to collect stool samples[15-21].
Nevertheless, in this situation it is impossible to differentiate indigenous from transitory microbiota and to make a reliable quantitative study of anaerobic or microaerophylic microorganisms, as to determine the different prevalence in various segments of the LDT.
Another study utilizing samples collected during laparotomy[22] did not respect the patient’s physiological conditions and the quantitative studies are also impaired by different dilutions of the mucous at the time of sampling.
Biopsies through colonoscopy, on the other hand, can be considered an aggressive procedure, when performed in healthy patients. Nevertheless, few papers are available in this field[23-25] and no method of collecting mucous aseptically has been described. Lack of concrete and reliable data motivated us to develop a more suitable collecting method. A special catheter was thus designed for collecting mucous avoiding biopsies. It can be adapted to the colonoscope, allowing collection of a sufficient amount of mucous without dilution and contact with air. The procedure is safe for the patient as it does not determine lesions or any other damage to the mucosa.
In this way, Uno et al[26] in 1998 developed a new catheter protected by a distal rubber covering, which blocks the infiltration of intestinal content during its passage through the surgical channel of colonoscope. This catheter does not work satisfactory as the protective cover itself contaminates the needle as it is introduced, therefore, contaminating the aspirate.
This problem was solved in this study by designing a catheter with the features mentioned in the Method section with a disposable protective membrane, as proposed by one of the authors (PRAA), which is easily removed by the airflow (Figure 1).
The reliability of this method was confirmed by the uniformity of the results regarding identification of the flora. Nevertheless, samples were collected during colonoscopy and bowel preparation was performed previously.
The impact of this preparation on the microflora probably promotes a reduction of the concentration but does not interfere with the quality, or with the final composition of this microbiota.
Also it must be stressed that the samples were collected from the mucous area but not from the intestinal lumen. Other studies also demonstrated[27-29] that repopulation of the microbiota is approximately five hours after bowel preparation.
In other hand, up to the present time, a qualitative and quantitative study of the normal indigenous microbiota of the LDT has not been described, due to all the mentioned difficulties, collection methods and microbiological analysis.
This study revealed more than 36 genera in the LDT, including the prevalence and concentration of each genus at ach site of the LDT (Table 1).
Thirty-three microorganisms with a mean concentration of 105 CFU/mL were identified in the terminal ileum. Among these microorganisms the most constant and prevalent were Veillonella sp and Klebisiella sp. The latter had a statistically significant predominance in this region.
A mean concentration of 105 CFU/mL microorganisms was identified in the cecum with Veillonella sp being the most constant. A mean concentration of 105 CFU/mL microorganisms was identified in the ascending colon with Veillonella sp being the most recurrent. A mean concentration of 105 CFU/mL microorganisms was identified in the transverse colon, with Veillonella sp and Enterococcus sp being the most constant. A mean concentration of 105 CFU/mL microorganisms was identified in the descending colon, with Veillonella sp being the most frequent. A mean concentration of 105 CFU/mL microorganisms was identified in the sigmoid colon, with Veillonella sp, Clostridium sp, Corynebacterium sp, E.coli, Klebisiella sp and Enterobacter sp being the most constant. The latter displayed a statistically significant dominance. A mean concentration of 105 CFU/mL microorganisms was identified in the rectum with Veillonella sp and E.coli the most prevalent and a statistically significant predominance.
Also the frequent participation of Veillonella sp (mean concentration 105 CFU/mL) was observed at all sites examined, showing that there is a low oxidation-reduction (redox) potential in the LDT, since this bacterium lives only in an anaerobiotic environment (low redox).
It must be also stressed that some results and con-ventional believes have not been proved in this study, for example the very low concentration and prevalence of Lactobacillus sp at all sites. These results might be explained by the method of sampling using aspirated mucous instead of stools[29].
Moreover, Lactobacillus sp was only found in the rectum, and Candida albicans were occasionally found in healthy volunteers.
Also there was a similar distribution of aerobics and anaerobic microorganisms, demonstrating that there is no predominance of anaerobic specimens in the mucous but a similarity between aerobic and anaerobic bacteria although the redox potential was low.
A higher prevalence of the bacteria was found in the rectum using this standardized sampling method.
The presence of Veillonella sp, Klebisiella sp, Clostridium sp, E.coli and Corynebacterium sp was observed in specimens from normal volunteers and could be considered as a biological marker. In addition, Bacillus sp, Bifidobacterium sp, Candida sp, Eubacterium sp, Fusobacterium sp, Peptococcus sp, Peptostreptococcus sp, Propionibacterium sp, Proteus sp, Pseudomonas sp, Rodothorula sp, Selemonas sp, Staphylococcus sp, Streptococcus sp practically were not found in the healthy LDT, suggesting that their identification in significant concentration could indicate a pathological status.
In conclusion, our sampling method is efficient for obtaining suitable samples of mucous from the LDT for qualitative and quantitative microbiological studies. This methodology creates perspectives for studying and determining new criteria and concepts as well as for standardization of future prophylactive treatment[30] in gastroenterology.
S- Editor Zhu LH L- Editor Wang XL E- Editor Liu Y
| 1. | Symposium: Status and perspectives in gastro-intestinal microecology. Potsdam, GDR, 24-26 April 1984. Proceedings. Nahrung. 1987;31:355-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 2. | van der Waaij D, Berghuis-de Vries JM. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond). 1971;69:405-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 606] [Cited by in RCA: 544] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 3. | Freter R. Interactions between mechanisms controlling the intestinal microflora. Am J Clin Nutr. 1974;27:1409-1416. [PubMed] |
| 4. | Alves PRA, Souza AHS, Habr-Gama A, Gama-Rodrigues JJ, Pinotti HW. Express mannitol: a safe and fast bowel preparation for colonoscopy used on 3 400 consecutive patients. ABCD Arq Bras Cir Dig. 1991;6:20-23. |
| 5. | Quintanilha AG, Zilberstein B, Santos MAA, Pajecki D, Moura EG, Martins B, Gama-Rodrigues JJ. Sample method for determination of the lower gastrointestinal tract microbiota. In: 13th World Congress of the International Association of Surgeons and Gastroenterologists. Hepato-Gastroenterology. 2003;50 suppl 1:161. |
| 6. | Zilberstein B. Quintanilha AG, Santos MAA, Pajecki D, Alves PRA, Maluf F, Gama-Rodrigues JJ. Qualitative and quantitative investigation healthy volunteer lower gastrointestinal tract microbiota. 13th World Congress of the International Association of Surgeons and Gastroenterologists. Hepato-Gastroenterology. 2003;50 Suppl 1:160. |
| 7. | Möller AJ. Microbiological examination of root canals and periapical tissues of human teeth. Methodological studies. Odontol Tidskr. 1966;74 Suppl:1-380. [PubMed] |
| 8. | Krieg NR, Holt JG. Bergey's manual of systematic bacteriology. London: Willians & Wilkins 1992; . |
| 9. | Siegel S. Estatística não paramétrica: para as ciências do comportamento. São Paulo: MacGraw Hill do Brasil 1975; 75. |
| 10. | Ma LS, Pan BR. Strengthen international academic exchange and promote development of gastroenterology. World J Gastroenterol. 1998;4:1. [PubMed] |
| 11. | Lam SK. Academic investigator's perspectives of medical treatment for peptic ulcer. Ulcer disease: investigation and basis for therapy. New York: Marcel Dekker 1991; 431-450. |
| 12. | Shiner M. A capsule for obtaining sterile samples of gastrointestinal fluids. Lancet. 1963;1:532-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | Kalser MH, Cohen R, Arteaga I, Yawn E, Mayoral L, Hoffert W, Frazier D. Normal viral and bacterial flora of the human small and large intestine. N Engl J Med. 1966;274:558-563 contd. [PubMed] |
| 14. | Belov L, Meher-Homji V, Putaswamy V, Miller R. Western blot analysis of bile or intestinal fluid from patients with septic shock or systemic inflammatory response syndrome, using antibodies to TNF-alpha, IL-1 alpha and IL-1 beta. Immunol Cell Biol. 1999;77:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Finegold SM, Attebery HR, Sutter VL. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456-1469. [PubMed] |
| 16. | Reddy BS, Wynder EL. Large-bowel carcinogenesis: fecal constituents of populations with diverse incidence rates of colon cancer. J Natl Cancer Inst. 1973;50:1437-1442. [PubMed] |
| 17. | Reddy BS, Weisburger JH, Wynder EL. Effects of high risk and low risk diets for colon carcinogenesis on fecal microflora and steroids in man. J Nutr. 1975;105:878-884. [PubMed] |
| 18. | Gustafsson BE. The physiological importance of the colonic microflora. Scand J Gastroenterol Suppl. 1982;77:117-131. [PubMed] |
| 19. | Bertazzoni EM, Benoni G, Berti T, Deganello A, Zoppi G, Gaburro D. A simplified method for the evaluation of human faecal flora in clinical practice. Helv Paediatr Acta. 1978;32:471-478. [PubMed] |
| 20. | Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34 Suppl 2:S12-S18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 176] [Article Influence: 7.7] [Reference Citation Analysis (1)] |
| 21. | Kaur IP, Chopra K, Saini A. Probiotics: potential pharmaceutical applications. Eur J Pharm Sci. 2002;15:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 175] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 22. | Bentley DW, Nichols RL, Condon RE, Gorbach SL. The microflora of the human ileum and intrabdominal colon: results of direct needle aspiration at surgery and evaluation of the technique. J Lab Clin Med. 1972;79:421-429. [PubMed] |
| 23. | Kuisma J, Mentula S, Luukkonen P, Jarvinen H, Kahri A, Farkkila M. Factors associated with ileal mucosal morphology and inflammation in patients with ileal pouch-anal anastomosis for ulcerative colitis. Dis Colon Rectum. 2003;46:1476-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 958] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 25. | Schiffrin EJ, Blum S. Interactions between the microbiota and the intestinal mucosa. Eur J Clin Nutr. 2002;56 Suppl 3:S60-S64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Uno Y, Munakata A, Ohtomo Y. Farewell to bacteremia caused by endoscopic injection--effectiveness of a new injection catheter with a covered tip. Gastrointest Endosc. 1998;47:523-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 27. | Arabi Y, Dimock F, Burdon DW, Alexander-Williams J, Keighley MR. Influence of bowel preparation and antimicrobials on colonic microflora. Br J Surg. 1978;65:555-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Nichols RL, Condon RE. Preoperative preparation of the colon. Surg Gynecol Obstet. 1971;132:323-337. [PubMed] |
| 30. | Zilberstein B, Cleva R, Quintanilha AG. A Microbiota do trato gastrointestinal e as infecções em cirurgia do aparelho digestivo. ABCD Arq Bras Cir Dig. 1995;10:91-92. |