Published online Jul 21, 2007. doi: 10.3748/wjg.v13.i27.3747
Revised: April 10, 2007
Accepted: April 18, 2007
Published online: July 21, 2007
AIM: To establish the role of human T Cell Factor-4 (hTCF-4) gene exons 3-9 mutation status in association with sporadic rectal cancer with microsatellite instability (MSI).
METHODS: Microsatellite markers were genotyped in 93 sporadic rectal cancer patients. Eleven cases were found to be high-frequency MSI (MSI-H). Sequence analysis of the coding region of the exons 3-9 of hTCF-4 gene was carried out for the 11 MSI-H cases and 10 controls (5 microsatellite stability (MSS) cases and 5 cases with normal mucosa). The sequencing and MSI identification were used.
RESULTS: Several novel mutations and variants were revealed. In exon 4, one is a 4-position continuous alteration which caused amino acid change from Q131T and S132I (391insA, 392 G > A, 393 A > G and 395delC) and another nucleotide deletion (395delC) is present in MSI-H cases (5/10 and 4/10, respectively) but completely absent in the controls.
CONCLUSION: Novel mutations in exon 4 of hTCF-4 gene were revealed in this study, which might be of importance in the pathogenesis of sporadic rectal cancer patients with MSI-H.
- Citation: Meng WJ, Wang L, Tian C, Yu YY, Zhou B, Gu J, Xia QJ, Sun XF, Li Y, Wang R, Zheng XL, Zhou ZG. Novel mutations and sequence variants in exons 3-9 of human T Cell Factor-4 gene in sporadic rectal cancer patients stratified by microsatellite instability. World J Gastroenterol 2007; 13(27): 3747-3751
- URL: https://www.wjgnet.com/1007-9327/full/v13/i27/3747.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i27.3747
Colorectal cancer is the second leading cause of cancer related deaths in the Western world. It is the third leading cause of cancer deaths and tends to increase in China. The mechanism of the malignant transformation of colorectal cells is far from being clearly understood. Involvement of the Wnt signaling pathway, also called the APC/β-catenin/TCF pathway, has been demonstrated notably in colorectal cancers. Wnts act by stabilizing cellular levels of the transcriptional coactivator β-catenin, which forms complexes with sequence-specific DNA binding T cell factor/lymphocyte enhanced factor (TCF/LEF) transcription factors. In the absence of nuclear β-catenin, TCF/LEFs act as transcriptional repressors by recruiting proteins such as Groucho/Transducin-like enhancer-of-split (TLE), and COOH-terminal-binding protein (CtBP). Upon Wnt activation, the binding of β-catenin to TCFs generates a bipartite transcription factor, in which TCFs provides the DNA binding domain and the C terminus of β-catenin provides the transactivation domain, therefore inducing a transcriptional switch. However, the molecular basis of the switch from transcriptional repression to activation during Wnt signaling is still not clear.
TCF-4 (also known as TCF7L2) is a sequence-specific transcriptional factor, whose functions of the downstream effectors of Wnt/β-catenin signals are similar to the other human TCF proteins (TCF-1, TCF-2/LEF-1, and TCF-3) TCF-4 is the most intensively expressed member of the TCF/LEF gene family in normal colonic tissues. In the TCF-4 gene, three major domains are present: β-catenin-binding domain in exon 1, the DNA-binding HMG boxes in exon 10 and 11, and the COOH-terminal-binding domain in exon 17[1,2]. The human TCF-4 (hTCF-4) gene was reported to be one of the targets of microsatellite instability (MSI) in colorectal cancers. Recently, in colorectal cancer with high-frequency MSI (MSI-H), frequent frameshift mutations involved in hTCF-4 exon 17 have been found to modulate transcriptional activity through the truncation at the COOH-terminal region[3,4]. However, Ruckert et al[5] found that mutations in the A9 repeat of hTCF-4 exon 17 do not contribute to tumorigenesis. The hTCF-4 protein has been shown to repress transcription by recruiting corepressor proteins not only CtBP, but also Groucho/TLE[6]. A major advance in explaining how TCFs repress transcription followed the discovery that Groucho/TLE is a specific binding partner of TCFs. Binding occurs between the Q-domain of Groucho and a conserved region in TCFs between the β-catenin binding domain and the HMG box[7]. The conserved region was encoded by exons 3-9. Therefore, whether mutations of the region interfere with binding to Groucho/TLE family proteins remains to be elucidated. To gain insight into the molecular basis of rectal cancer, we have screened hTCF-4 mutations in human rectal tumors in exons 3-9. The up-to-date methods of sequencing and MSI identification were used. The data showed that novel mutations in exon 4 of hTCF-4 gene might be of importance in the pathogenesis of sporadic rectal cancer patients with MSI-H.
One hundred and two patients from Chinese Han population who underwent surgical resection for sporadic rectal cancer at West China Hospital of Sichuan University (Sichuan, China) from 2002 to 2005 were included in the study. All cases were deemed sporadic, based on the absence of relevant family histories as recorded prospectively at initial patient interview. Patients who were suspected clinically to have HNPCC fulfilling the Amsterdam I/II[8] or Bethesda criteria[9] were excluded. Patients treated by preoperative radiotherapy or chemotherapy were also excluded. Of 102 cases, 9 were excluded because of tissue unavailability, resulting in a total of 93 eligible cases. Tumor histotype and grading were defined according to the recommendations of the World Health Organization for the histological typing of colorectal cancer[10]. All cases were staged using the 6th edition of the American Joint Committee on Cancer (AJCC) Staging Manual[11]. Informed consent was provided by all subjects, and approval was obtained from the Regional Ethics Committee.
Samples of tumor tissues and of normal mucosa were frozen immediately after surgery in liquid nitrogen and stored at -80°C until analysis. Before DNA extraction, the presence of adequate neoplastic material (at least 60%-70% of tumor cells) was verified by microscopic examination. Genomic DNAs of tumor and corresponding normal mucosa were extracted by a standard phenol-chloroform procedure. Total RNA was extracted from each subject using Trizol (Invitrogen Co., Ltd) method. Total RNA of each sample was dissolved in RNase-free water and stored in the refrigerator at -80°C before use. The concentration of DNA and RNA was determined by spectrophotometer, and their integrity was checked by each gel electrophoresis.
Primers used to amplify simple sequence repeat markers were obtained from Life Technologies (TaKaRa, Dalian, China). MSI was studied at five loci recommended by a National Cancer Institute workshop on MSI (BAT25, BAT26, D2S123, D5S346 and D17S250)[12]. All primer sequences were as reported in Genome DataBase (GDB; http://www.gdb.org). Lesions were characterized as MSI-H if they manifested instability at two or more of the five loci in tumor DNA, and as microsatellite stability (MSS) if showing no instability at any loci when compared with normal DNA. For each marker analysis, PCR was carried out as previously described[13]. Electrophoreses of the amplified PCR products were performed in 2% agarose gels containing goldview in Tris-Borate EDTA buffer. Gels were examined using an UV transilluminator and verified as a single band. Electrophoresis of PCR products was separated on a 6% denaturing polyacrylamide gel containing 8 M urea. Gels were stained with silver salts according to Sanguinetti et al[14]. Experiments to ascertain MSI were done independently two to three times for each genetic locus. Through detection of MSI, 11 cancers demonstrating MSI-H and 10 controls (5 MSS and 5 with normal mucosa) were included for mutation analysis.
Screening for hTCF-4 exons 3-9 mutations was performed by RT-PCR of total RNA. Two-step methods were adopted. Reverse transcription conditions for all PCRs were optimized on iCycler iQ (Bio-Rad, USA) and the optimum annealing temperature was 53.1°C. The following iCycler iQ run protocol was used: denaturation program (95°C, 5 min), amplification programs repeated 50 times (95°C for 20 s, 53.1°C for 30 s and 72°C for 30 s). The first half of the exons 3-9 was amplified using F1 (5’CGCCAACGACGAACTGAT3’) and R1 (5’GCACCACTGGCACTTTGT3’) primers, whereas the second half was amplified using F2 (5’ATGAAATGGCCACTGCTTGA3’) and R2 (5’CCTTTTTGGAGTCCTGATGCT3’) primers. The annealing temperature was 56°C and 58°C, respectively. Presence of all sequence variants was confirmed by performing three independent PCR reactions and subsequent DNA sequencing. In addition, amplification for hTCF-4 exon 4 was performed by PCR of genomic DNA. The primers and conditions were used according to Duval et al[15]. For PCR reaction, a tube without template DNA served as a negative control.
Electrophoreses of the amplified products were performed in 2% agarose gels containing goldview in Tris-Borate EDTA buffer. Amplicons were purified by solution extraction and bidirectional sequencing using an ABI Prism 377 DNA sequencer.
To determine the potential deleterious effect of the amino acid changes, we used SIFT (http://blocks.fhcrc. org/sift/SIFT.html). The SIFT software uses the protein sequence similarity of different species and the hydrophobic characteristics of amino acids to calculate the probability of the deleterious effect of specific amino acid variants[16]. Scores lower than 0.05 suggest a potential pathogenic amino acid substitution.
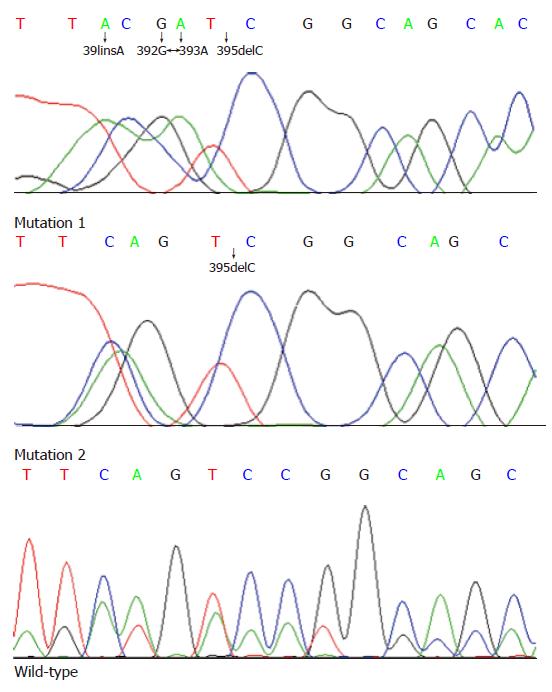
A total of 11 cases demonstrated MSI-H using markers recommended by a National Cancer Institute workshop by PCR amplification from 93 sporadic rectal cancer patients in our study. Sequencing data collection and analysis were successfully performed for the hTCF-4 gene (exons 3-9) in these MSI-H cases (n = 10) and controls (n = 10) except for one MSI-H case. This study revealed several novel mutations and sequence variants between exons 3-9. The sequence at the beginning of exon 4 showed a TACGATCG repeat, which was present in 5 of MSI-H cases but not in the controls, as shown in Figure 1 and Table 1. Sequencing of exon 4 revealed a deletion of cytosine at 395 (395delC) which was only found in 4 MSI-H cases without above-mentioned mutation. Genetic and clinical details of these 4 cases are given in Tables 1 and 2, respectively.
| Frequency | |||||
| Exon | Nucleotidechange | Amino acidchange | Mutationtype | Patients(n = 10) | Controls(n = 10) |
| 4 | 4-position | ||||
| continuous | |||||
| alteration1 | Q131T | Missense | 5 | 0 | |
| 4 | 395delC | S132I | Frameshift | 4 | 0 |
| 4 | 450 G > C | Q150H | Missense | 1 | 0 |
| 8 | 868 G > A | V290M | Missense | 2 | 0 |
| Case | Age | Sex | Size | Pathology | TNM | CEA |
| No. | (yr) | (mm) | stage | (ng/mL) | ||
| 1 | 54 | F | 20 | Mucinous | I | 1.55 |
| 2 | 28 | F | 50 | Mucinous | IV | 310.5 |
| 3 | 68 | F | 60 | Mucinous | II | 3.47 |
| 4 | 49 | F | 40 | Mucinous | II | 1.6 |
| 5 | 51 | M | 40 | Unmucinous | II | 10.11 |
| 6 | 36 | M | 100 | Mucinous | II | 6.37 |
| 7 | 71 | F | 40 | Unmucinous | I | 3.53 |
| 8 | 47 | M | 40 | Unmucinous | III | 4.96 |
| 9 | 54 | M | 80 | Unmucinous | II | 4.3 |
| 10 | 54 | M | 80 | Unmucinous | III | 5.8 |
In addition, there was a missense mutation (450 G > C) in exon 4 in one MSI-H case, resulting in transition of Glutamine to Histidine (Q150H) when translated. Finally, a change of 868 G to A, leading to a V290M amino acid change, was observed in exon 8 in two MSI-H cases. These novel variants were not present in any of the controls either. SIFT software suggested no potential deleterious effect of the two amino acid changes. No mutation was observed in other exons. The mutation (C to T) at the nucleotide 35 of exon 4 detected by DGGE only in the SW48 cell line was not observed in our study[15].
A link was previously established between hTCFs and Wnt signaling, a pathway that plays a crucial role in many developmental processes as well as in human carcinogenesis[1]. Although it is well established that the formation of nuclear β-catenin/TCF complexes plays a pivotal role in the activation of Wnt target genes, the exact mechanisms of transcriptional activation and regulation are still under investigation[17,18]. Duval et al[2] reported frequent frameshift alterations in an A9 coding repeat localized in exon 17 of hTCF-4 in MSI-H colorectal cancers and the main consequence of such a mutation was to change hTCF-4 transactivating properties by modifying the respective proportions of the different isoforms containing CtBP binding domains. However, Ruckert et al[5] found that the mutations do not contribute to tumorigenesis. Thus, the question is if mutations of the Groucho/TLE binding domain encoded by exons 3-9 interfere with binding to Groucho/TLE family proteins and remove the repressive effect of Groucho/TLE proteins. To our knowledge, there has been no report on it.
Sequencing data collection and analysis were successfully performed for the hTCF-4 gene (exons 3-9) in these MSI-H cases (n = 10) and controls (n = 10) except for one MSI-H case. This study revealed several novel mutations and sequence variants between exons 3-9. The sequence at the beginning of exon 4 showed a TACGATCG repeat which did not match perfectly to the TCAGTCCG repeat in the previously published hTCF-4 mRNA sequence[15]. Although an explanation regarding the apparent discrepancy is not forthcoming, the determinacy of this sequence variant may be supported by the following: Firstly, the sequence was confirmed by repetition of three independent PCR and sequencing reactions, including sequencing in reverse direction. Secondly, the sequence variant was present in 5 MSI-H cases but not in the controls. Sequence alignments have shown that although the 4-position continuous alteration (391insA, 392 G > A, 393 A > G and 395delC) did not alter the whole reading frame, the change from a Serine to a highly hydrophobic Isoleucine (S132I) is likely to have any functional relevance.
Another finding in this study was that a novel mutation could be implicated in the rectal cancer pathogenesis. Sequencing of exon 4 revealed a deletion of cytosine at 395 (395delC) only in 4 MSI-H cases without above-mentioned mutation. The absence of this 394delC in the controls suggests a potential pathogenic effect. The 394delC altered reading frame and 22 amino-acid peptides are encoded instead of a full-length protein. Therefore, the mutation in hTCF-4 exon 4 may modulate the switch status through the truncation between the β-catenin binding domain and the HMG box DNA-binding region (Groucho/TLE binding domain) of the hTCF-4 protein, which suggests this mechanism could be of functional significance for regulating hTCF-4 transcriptional activity. Except for this mutation associated with female and mucinous caricinoma patients (P < 0.05) (data not shown), no other clinicopathological features (such as age at diagnosis, tumor size, TNM stage, level of preoperative serum CEA and differentiation) could be identified in these 4 patients associated with mutation 395delC. We infer that the upstream of exon 4 is an unstable area so that it is subjected to mutations, which may be one of the mechanisms of the tendency to be truncated in colorectal tumors. This indicates that these novel mutations in exon 4 of hTCF-4 gene revealed in the study, might be of importance in the pathogenesis of sporadic rectal cancer patients with MSI-H.
In conclusion, this study provides important evidence for novel mutations and sequence variants in association with the pathogenesis of sporadic rectal cancer with MSI-H. These sequence variants in Chinese Han population indicates the diversity of human library and the complexity of rectal carcinogenesis. Further confirmatory studies on the spectrum, prevalence rates, and functional effect of sequence variants in the exons 3-9 of hTCF-4 gene, in particular exon 4, in other populations should further demonstrate the true contribution of this gene to colorectal carcinogenesis.
We would thank Dr. JH Sui of Harvard University for the critical reading of the manuscript.
A link was previously established between hTCFs and Wnt signaling which plays a crucial role in many developmental processes as well as in human carcinogenesis. Although it is well established that the formation of nuclear β-catenin/TCF complexes plays a pivotal role in the activation of Wnt target genes, the exact mechanisms of transcriptional activation and regulation are still under investigation. Recently, in colorectal cancer with MSI-H, frequent frameshift mutations involved in hTCF-4 exon 17 have been reported to modulate transcriptional activity through the truncation at the COOH-terminal region. However, Ruckert et al. found that the mutations do not contribute to tumorigenesis. Thus, the question is if mutations of the Groucho/TLE binding domain encoded by exons 3-9 interfere with binding to Groucho/TLE family proteins and remove the repressive effect of Groucho/TLE proteins. We have screened for hTCF-4 mutations in human rectal tumors in exons 3-9.
The mechanism of colorectal cancer is far from being clearly understood. Involvement of the Wnt signaling pathway has been demonstrated notably in colorectal cancers. Wnts act by stabilizing cellular levels of the transcriptional coactivator β-catenin, which forms complexes with sequence-specific DNA binding TCF/LEF transcription factors. However, the molecular basis of the switch from transcriptional repression to activation during Wnt signaling is not clear. TCF-4 is sequence-specific HMG box transcriptional factors that function as the downstream effectors of Wnt/β-catenin signals. TCF-4 is the most intensively expressed member of the TCF/LEF gene family in normal colonic tissue. The hTCF-4 gene was reported to be one of the targets of MSI in colorectal cancers. Recently, in colorectal cancer with MSI-H, frequent frameshift mutations involved in hTCF-4 exon 17 have been found to modulate transcriptional activity through the truncation at the COOH-terminal region. However, Ruckert et al found that mutations in the A9 repeat of hTCF-4 exon 17 do not contribute to tumorigenesis. The hTCF-4 protein has been shown to repress transcription by recruiting corepressor proteins not only CtBP, but also Groucho/TLE. A major advance in explaining how TCFs repress transcription followed the discovery that Groucho/TLE is a specific binding partner of TCFs. Binding occurs in a conserved region in TCFs encoded by exons 3-9. Therefore, whether mutations of the region interfere with binding to Groucho/TLE family proteins remain to be elucidated.
The mutation in hTCF-4 exon 4 may modulate the switch status through the truncation Groucho/TLE binding domain of the hTCF-4 protein, which suggest this mechanism could be of functional significance for regulating hTCF-4 transcriptional activity. We infer that the upstream of exon 4 is an unstable area so that it is subjected to mutations, which may be one of the mechanisms of the tendency to be truncated in colorectal tumors. This indicates that these novel mutations in exon 4 of hTCF-4 gene revealed in the study, might be of importance in the pathogenesis of sporadic rectal cancer patients with MSI-H.
This present study provides an important evidence for novel mutations and sequence variants in association with the pathogenesis of sporadic rectal cancer with MSI-H. These sequence variants in Chinese Han population indicate the diversity of human library and the complexity of rectal carcinogensis.
This is an interesting study investigating the possible association between TCF-4 exon 3-9 mutations and MSI high rectal cancer. It is a study reporting on previously uninvestigated area. The main conclusion of the authors was that new mutations discovered in the study might be of importance in the pathogenesis of sporadic rectal cancer patients with MSI-H.
S- Editor Liu Y L- Editor Ma JY E- Editor Lu W
| 1. | Clevers H, van de Wetering M. TCF/LEF factor earn their wings. Trends Genet. 1997;13:485-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 223] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 2. | Duval A, Gayet J, Zhou XP, Iacopetta B, Thomas G, Hamelin R. Frequent frameshift mutations of the TCF-4 gene in colorectal cancers with microsatellite instability. Cancer Res. 1999;59:4213-4215. [PubMed] |
| 3. | Duval A, Iacopetta B, Ranzani GN, Lothe RA, Thomas G, Hamelin R. Variable mutation frequencies in coding repeats of TCF-4 and other target genes in colon, gastric and endometrial carcinoma showing microsatellite instability. Oncogene. 1999;18:6806-6809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Saeki H, Tanaka S, Tokunaga E, Kawaguchi H, Ikeda Y, Maehara Y, Sugimachi K. Genetic alterations in the human Tcf-4 gene in Japanese patients with sporadic gastrointestinal cancers with microsatellite instability. Oncology. 2001;61:156-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 5. | Ruckert S, Hiendlmeyer E, Brueckl WM, Oswald U, Beyser K, Dietmaier W, Haynl A, Koch C, Rüschoff J, Brabletz T. T-cell factor-4 frameshift mutations occur frequently in human microsatellite instability-high colorectal carcinomas but do not contribute to carcinogenesis. Cancer Res. 2002;62:3009-3013. [PubMed] |
| 6. | Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 293] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 7. | Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 529] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 8. | Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1688] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 9. | Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2154] [Cited by in RCA: 2215] [Article Influence: 105.5] [Reference Citation Analysis (1)] |
| 10. | Jass JR, Sobin LH. World Health Organization. International Histological Classification of Tumors. Histological Typing of Intestinal Tumors (ed2)., Berlin, Germany, Springer-Verlag. 1989;. [DOI] [Full Text] |
| 11. | Greene FL, Page DL, Fleming ID, Fritz AG, Balch CM, Haller DG, Morrow M. AJCC Cancer Staging Manual, 6th ed, New York, Springer-Verlag. 2002;. [DOI] [Full Text] |
| 12. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [PubMed] |
| 13. | Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545-4549. [PubMed] |
| 14. | Sanguinetti CJ, Dias Neto E, Simpson AJ. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques. 1994;17:914-921. [PubMed] |
| 15. | Duval A, Rolland S, Tubacher E, Bui H, Thomas G, Hamelin R. The human T-cell transcription factor-4 gene: structure, extensive characterization of alternative splicings, and mutational analysis in colorectal cancer cell lines. Cancer Res. 2000;60:3872-3879. [PubMed] |
| 16. | Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812-3814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4033] [Cited by in RCA: 4463] [Article Influence: 202.9] [Reference Citation Analysis (0)] |
| 17. | Bienz M, Clevers H. Armadillo/beta-catenin signals in the nucleus--proof beyond a reasonable doubt? Nat Cell Biol. 2003;5:179-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 111] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Hecht A, Kemler R. Curbing the nuclear activities of beta-catenin. Control over Wnt target gene expression. EMBO Rep. 2000;1:24-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 135] [Article Influence: 5.4] [Reference Citation Analysis (0)] |