Published online May 28, 2007. doi: 10.3748/wjg.v13.i20.2791
Revised: February 10, 2007
Accepted: February 24, 2007
Published online: May 28, 2007
AIM: To investigate the association between the configurational and compositional changes of nuclear matrix and the differentiation of carcinoma cells.
METHODS: Cells cultured with or without 5 × 10-3 mmol/L of hexamethylene bisacetamide (HMBA) on Nickel grids were treated by selective extraction and prepared for whole mount observation under electron microscopy. The samples were examined under transmission electron microscope. Nuclear matrix proteins were selectively extracted and subjected to subcellular proteomics study. The protein expression patterns were analyzed by PDQuest software. Spots of differentially expressed nuclear matrix proteins were excised and subjected to in situ digestion with trypsin. The peptides were analyzed by matrix-assisted laser-desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS). Data were submitted for database searching using Mascot tool (www.matrixscience.com).
RESULTS: The nuclear matrix (NM) and intermediate filament (IF) in SMMC-7721 hepatocarcinoma cells were found relatively sparse and arranged irregularly. The nuclear lamina was non-uniform, and two kinds of filaments were not tightly connected. After induction for differentiation by HMBA, the NM-IF filaments were concentrated and distributed uniformly. The heterogeneous population of filaments, including highly branched utrathin filaments could also be seen in the regular meshwork. The connection between the two kinds of filaments and the relatively thin, condensed and sharply demarcated lamina composed of intermediate-sized filaments was relatively fastened. Meanwhile, 21 NM proteins changed remarkably during SMMC-7721 cell differentiation. Four proteins, i.e. mutant Pyst1, hypothetical protein, nucleophosmin1, and LBP were downregulated, whereas four other proteins, eIF6, p44 subunit, β-tubulin, and SIN3B were upregulated with the last one, SR2/ASF found only in the differentiated SMMC-7721 cells.
CONCLUSION: The induced differentiation of SMMC-7721 cells by HMBA is accompanied by the configurational changes of nuclear matrix-intermediate filament (NM-IF) system and the compositional changes of nuclear matrix protein expression. These changes may be important morphological or functional indications of the cancer cell reversion.
- Citation: Tang J, Niu JW, Xu DH, Li ZX, Li QF, Chen JA. Alteration of nuclear matrix-intermediate filament system and differential expression of nuclear matrix proteins during human hepatocarcinoma cell differentiation. World J Gastroenterol 2007; 13(20): 2791-2797
- URL: https://www.wjgnet.com/1007-9327/full/v13/i20/2791.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i20.2791
The nuclear matrix not only plays an important role in the process of DNA replication, hRNA processing and steroid hormone action, but also has direct effect on cell division and proliferation through constructing the architecture of higher order chromatin[1-3]. Moreover, it is often the important binding site of expression and regulation of oncogenes[4,5]. Previous researches have proved that the ultrastructural change of cytoskeleton and nuclear matrix was closely related to cell proliferation and differentiation[6]. The nuclear matrix or nuclear matrix associatied proteins have strong effects on the regulation of signal transduction, mRNA modification or translation, and more important, on the expression of oncogenes or tumor suppressor genes such as rb, p21, p53, which is extremely significant in the cell cycle processing and cell differentiation[7-10]. The nuclear matrix in cancer cells is not only abnormal in morphology, but also apparently different in its composition[12]. Our early studies showed distinct changes of the morphology or protein composition of nuclear matrix during differentiation of several cell lines treated with different reagents[12,15]. It is implied that those differentially expressed nuclear matrix proteins not only have direct association with regulators of cell cycle and signal transduction, but also are important during the cancerization and its reversion of the cell. Based on our former research of the effect on differentiation of SMMC-7721 cells with HMBA inducement[14], we studied the features and protein alternations of the nuclear matrix-intermediate filament (NM-IF) system in human hepatocarcinoma cell line SMMC-7721 during its differentiation induced by hexamethylamine bisacetamide (HMBA). It is helpful for further exploration and illustrating the relationship between the nuclear matrix and the cancerization and malignant phenotypic reversion of cancer cells.
HMBA (Sigma Chemical Company) was used to induce the differentiation of SMMC-7721 cells. Sequence grade, modified trypsin (Promega) and iodoacetamide (Sigma) were used in the in-gel digestion. ReadyStrip IPG strips (pH 3-10, 11 cm) and IPG buffer pH 3-10 were obtained from Amersham Biosciences. Other reagents used in 2-D gel electrophoresis and Coomassie blue R250 were from Shanghai Sangon Biological Engineering Technology and Service Co., Ltd.
SMMC-7721 human hepatocarcinoma cells were cultured at 37°C in RPMI-1640 medium (pH 7.2) supplemented with 10% newborn calf serum, 100 U/mL penicillin, 100 U/mL streptomycin and 50 μg/mL kanamycin. Cells of experimental group were cultured at the same medium added with 5 × 10-3 mol/L HMBA (Sigma). SMMC-7721 cells and the treated cells were seeded in small culture flasks with cover slip strips on which some nickel grids were covered with formvar and carbon film was sticked with polylysine, and grown in the normal medium and the medium containing 5 × 10-3 mol/L HMBA respectively. Fresh culture media were added to the cells every 48 h, and cells were harvested at subconfluency. The obtained cells were then stored at -80°C.
The cells were selectively extracted as described in our previous article[12]. Cover slip strips were removed from the culture medium and rinsed in phosphate-buffered saline at 4°C. Then they were placed in high ionic strength extraction medium [10 mmol/L PIPES (pH 6.8), 250 mmol/L (NH4)2SO4, 300 mmol/L sucrose, 3 mmol/L MgCl2, 1.2 mmol/L PMSF, 0.5% Triton X-100; CSK-AS] 4°C for 3 min. The extracted cells were rinsed briefly in digestion medium [10 mmol/L PIPES (pH 6.8), 50 mmol/L NaCl, 300 mmol/L sucrose, 3 mmol/L MgCl2, 1.2 mmol/L PMSF, 0.5% Triton X-100] without enzymes. Grids with anchored skeletons were placed in the digestion medium and appropriate enzymes (400 mg/L DNase I and 400 mg/L RNase A) and were incubated for 20 min at 23°C. The grids were then placed in high ionic strength extraction medium CSK-AS for 5 min at 23°C. Thus, only the nuclear matrix-intermediate filament structure remained intact.
The NM-IF samples on the grids after the selective extraction were prefixed in 2% glutaraldehyde at 4°C for 30 min, followed by 1% OsO4 in 0.1 mol/L sodium cacodylate (pH 7.2) for 5 min at 4°C. The cells, still attached to grids, were dehydrated in ethanol, dried through the CO2 critical point and examined under a JEM-100CXII/S transmission electron microscopy.
The SMMC-7721 cells were washed with PBS and extracted with cytoskeleton buffer (CSK100) (10 mmol/L PIPES pH 6.8, 300 mmol/L sucrose, 100 mmol/L NaCl, 4 mmol/L CaCl2, 1.0 mmol/L PMSF, 0.5% Triton X-100) at 0°C for 10 min, and subjected to centrifugation for 5 min at 400 r/min. The deposition was washed twice with CSK50 (10 mmol/L PIPES pH 6.8, 300 mmol/L sucrose, 50 mmol/L NaCl, 4 mmol/L CaCl2, 1.0 mmol/L PMSF, 0.5% Triton X-100) and digested for 30 min at 25°C in the same buffer containing 300 U/mL DNase I. One mole per liter ammonium sulfate was added dropwise to a final concentration of 0.25 mmol/L. After incubation for 15 min, the nuclear matrix proteins were pelleted by centrifugation at 1000 r/min for 5 min, and washed once with the CSK50 buffer, then stored at -80°C. Protein concentrations were determined by the method of Bradford.
To solubilize nuclear matrix proteins, the pellet of purified nuclei was resuspended in 2-D buffer containing 7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 50 mmol/L DTT and ultrasonicated for 2 min. The supernatant was centrifuged for 30 min at 16 000 ×g at 4°C. IEF was performed in ReadyStrip IPG strips. ReadyStrip IPG strips were rehydrated overnight in a reswelling tray with 2-D buffer containing 0.5% IPG buffer pH 3-10 and nuclear matrix proteins in a final volume of 250 μL (200 μg). IEF was carried out on a Protean IEF cell (Investigator) at 19°C with a maximum current setting of 80 mA/strip. Focusing was performed for a total of 70 000 V × h. Before carrying out the second dimensional SDS-PAGE, the strips were equilibrated in an equilibration buffer consisting of 20% glycerol, 2% SDS, 65 mmol/L Tris-HCl, pH 6.8 and 20 mmol/L DTT for 10 min at room temperature, then transferred into the second equilibration buffer containing 20% glycerol, 2% SDS, 65 mmol/L Tris-HCl, pH 6.8 and 2.5% iodoacetamide. The strips were transferred onto 1-mm thick SDS-PAGE gels and sealed in place with 1% agarose. SDS-PAGE was performed on a 12.5% acrylamide/bisacrylamide gel at 50 V for 30 min followed by 170 V for 8 h. The gels were run in the following electrode buffer: 25 mmol/L Tris, 192 mmol/L glycine, 0.1% SDS. SDS-PAGE standards were used for gel calibration.
The gel was fixed (50% methanol and 10% glacial acetic acid) for more than 40 min, sensitized (70 mL of methanol, 10 mL of 10% Sodium thiosulfate, 17 g of NaAC, bringing up to 250 mL with dH2O) for 30 min and rinsed in dH2O 3 times for 5 min each. The gel was stained in stain solution (25 mL of 5% AgNO3, 0.1 mL of formaldehyde, then increasing to 250 mL with dH2O) for 30 min rinsed in dH2O 3 times 1 min each and placed in developer (250 mL of dH2O, 6.25g Na2CO3, 50 μL of formaldehyde) until spots were almost as dark as desired. Development was stopped (3.75 g EDTA in 250 mL dH2O) and scanned using Magicscan. 2-DE maps of nuclear matrix proteins were subjected to analysis with PDQuest (Bio Rad) software.
Silver-stained spots were excised and washed with 50 mL fresh bleaching liquid (100 mmol/L Na2S2O3: 30 mmol/L K3Fe (CN)6 = 1:1). Gel spots were dried in a vacuum centrifuge and reswelled in 50 mL of solution containing 10 mol/L DTT/100 mol/L NH4HCO3 and incubated at 57°C for 1 h. This solution was subsequently replaced with 50 mL of solution containing 55 mol/L IAA, 100 mol/L NH4HCO3 and incubated at room temperature for 30 min. The gel spots were dried again and digested with TPCK-trypsin at 37°C overnight. After the incubation, the liquid was removed from the gel piece and the liquid was transferred to a new-labeled tube. This solution contains the extracted tryptic peptides.
For MALDI-TOF-MS analysis, samples were dissolved in 2 μL 0.1% TFA. Mass measurements were carried out on a Brucker ULTRAFLEXTM TOF/TOF mass spectrometer. This instrument was used at a maximum accelerating potential of 20 kV (in positive mode) and was operated in reflector mode. And 0.5 μL of saturated solution of α-cyano-4-hydroxy cinnamic acid in 0.1% TFA/30% acetonitrile was mixed with 0.5 μL sample solution, and added to the target. Internal calibration was performed with tryptic peptides coming from autodigestion of trypsin (monoisotopic masses at m/z 842.51, and m/z 2 211.10). Monoisotopic peptide masses were assigned and used for database search.
Peptide mass fingerprints obtained by the MALDI-TOF-MS were used to search nonredundant protein sequence database using Mascot software from Matrix Science. Search parameters included a maximum allowed peptide mass error of 100 ppm with consideration of one incomplete cleavage per peptide. Accepted modifications included carbamidomethylation of cysteine residues (from iodoacetamide exposure) and methionine oxidation, a common modification occurring during SDS-PAGE. The criteria for positive identification of proteins were set as follows: (1) Statistical significance (P < 0.05) of the match when tested by Mascot; (2) The matched peptides covered at least 17% of the whole protein sequence; (3) Concordance (± 15%) with the molecular weight and pI of the parent 2-D PAGE protein spot; and (4) Protein identifications not fulfilling criterion 2 were still assigned, if criteria 1 and 3 were fulfilled and no other homo-sapiens proteins with peptide mass-matched. P < 0.05 was identified by Mascot and identified protein wasinferred.
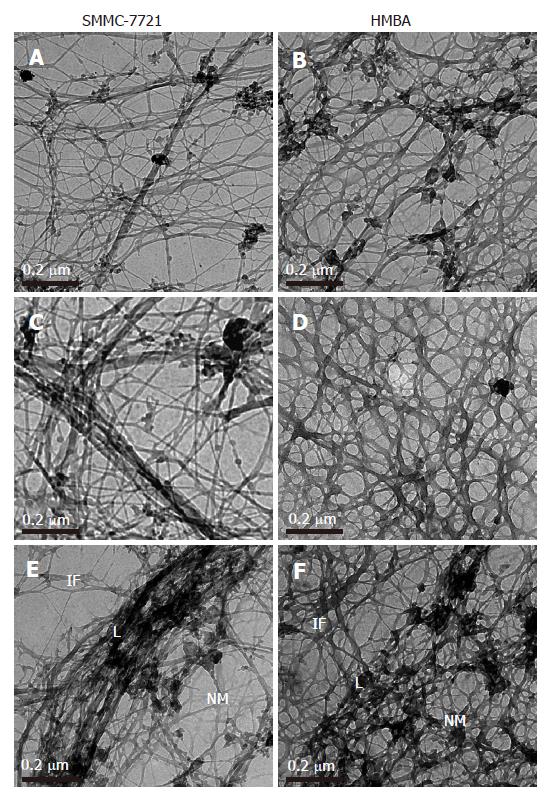
Selective extraction technique revealed cytoplasmic and nuclear meshworks in SMMC-7721 cells. The nuclear matrix was mainly composed of thick filaments. The peripheral region of the nucleus was demarcated by the wide band of densely packed filaments. This structure, representing nuclear lamina, was attached to the surroun-ding intermediate filaments (IF) of cytoskeleton. The nuclear matrix filaments in SMMC-7721 cells were relatively few and scattered, not well distributed and arranged irregularly within the nucleus region, and there were few single filaments while most of the nuclear matrix filaments were quite thick in bundle-like form and interweaved into an irregular meshwork (Figure 1A). The nuclear lamina was composed of densely packed fibrils, and it was ununiformly thick and compact. The inner nuclear lamina was connected with some thick nuclear matrix filament bundles or thin and short filaments. The intermediate filaments which terminated on the out nuclear lamina were few, always in thick bundles or in strip-rope-like structure, and arranged irregularly, too (Figure 1C and 1E). Nevertheless, in the SMMC-7721 cells induced by HMBA, the nuclear matrix filaments were abundant and well-distributed, different in slender and thick form in the nucleus region. The single filaments increased, and interweaved into a regular network (Figure 1B). The wide nuclear lamina turned into a thin and compact fibroid structure, the inner nuclear lamina was connected closely with the nuclear matrix filaments, and many long and slender intermediate filament bundles were terminated directly on the outer nuclear lamina. Both the nuclear matrix filaments and the intermediate filaments connected to the nuclear lamina increased and appeared quite dense. Moreover, the intermediate filaments in the treated cells were abundant and well-distributed in the cytoplasmic region (Figure 1D and 1F).
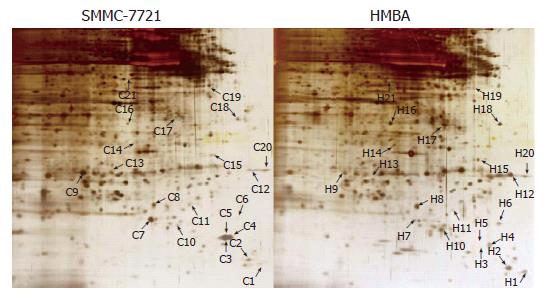
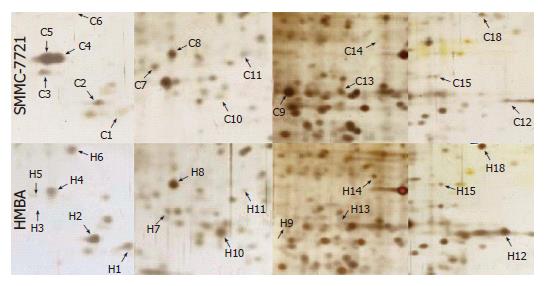
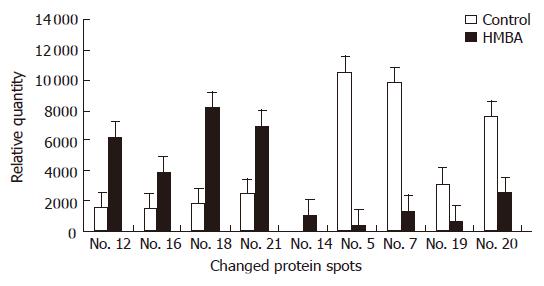
Samples of nuclear matrix proteins extracted from SMMC-7721 cells treated with or without 5 mmol/L HMBA were subjected to 2-D gel electrophoresis for at least three repeats per experimental condition. Analysis of proteins was based on evaluation of at least two gels. Twenty-one protein spots were changed remarkably, whereas most of the spots were similar to the control in the expression patterns of nuclear matrix proteins from differentiated SMMC-7721 cell. Among the changed spots, 8 spots (C4, C5, C7, C9, C11, C13, C19, C20) were downregulated, one spot disappeared in the differentiated SMMC-7721 cells, whereas 11 spots (C1, C2, C6, C8, C10, C12, C15, C16, C17, C18, C21, C3) were upregulated, and 1 spot (C14) emerged as a new protein spot in the differentiated cells (Figures 2 and 3). Relative expression levels of the changed proteins were shown using Melanie software (Figure 4), relative volume (% vol) of spot was employed to make data independent of uninteresting variation between gels, such as differences in protein loading or staining.
These 22 protein spots were cut out from the gels and analyzed with MALDI-TOF-MS. Peptide mass fingerprint (PMF) of each protein spot was then generated (data not shown). By searching the Mascot database, we identified 9 proteins combined with the searching results. The characteristics of the protein, the number and intensity of peptide matching peak, the sequence coverage of matching peptide, the theoretical and approximate values of Mr and pI, the identified protein names, accession numbers, the sequence coverages, and the theoretical Mr and pI values for each protein spot are listed in Table 1.
| Spot No. | NCBInr entry | Protein name | Mw/pI | Sequencecoverage(%) | Biological function |
| Up-regulated | |||||
| No. 12 | gi13785574 | p27BBP protein/eIF6 | 26 845/4.56 | 27 | β4 integrin interactor, a component of NM filaments escorts 60S ribosome subunit from nuclear to cytoplasm |
| No. 16 | gi88983724 | Similar to TFIIH | 36 539/6.79 | 23 | Basic transcriptin factor 2 44 kDa subunit (BTF2-p44) (General transcription factor) |
| Basal transcription factor complex p44 subunit | |||||
| No. 18 | gi14043898 | Tubulin, beta 2C | 50 255/4.79 | 17 | Tubulin β subunit, with GTPase activity |
| gi27368062 | Class IVb beta tubulin | 50 177/4.82 | 17 | ||
| No. 21 | gi13477279 | SIN3B protein | 49 609/6.56 | 23 | Interact with the Mad components of the Myc/Max/Mad network of cell growth regulators |
| New protein | |||||
| No. 14 | gi45708738 | SFRS1 protein | 22560/7.72 | 25 | arginine/serine-rich 1 alternate splicing factor |
| Down-regulated | |||||
| No. 5 | gi5822131 | Crystal structure of an active site mutant of the pyst1 | 16364/5.31 | 47 | ERK-specific MAP kinase phosphatase PYST1/MKP3 |
| No. 7 | gi62088338 | Hypothetical protein: DKFZp434K1815 variant | 26 864/5.64 | 24 | From Deutches Krebs-Forschungszentrum cancer research centre (German) |
| No. 19 | gi34234 | Laminin-binding protein | 31 888/4.84 | 28 | Laminin-binding protein, LBP |
| gi250127 | 67 kDa laminin receptor | 32 860/4.83 | 27 | ||
| gi17939541 | Integrin beta 4 binding protein | 27 095/4.56 | 26 | ||
| No. 20 | gi83641870 | Nucleophosmin1 | 28 497/4.56 | 23 | Also known as NPM, numatrin, No38, B23 protein |
| Isoform 3 | |||||
| gi40353734 | Nucleophosmin 1 | 29 617/4.47 | 23 | A major nucleolus phosphoprotein, a component of NM | |
| Isoform 2 |
The abnormality of NM-IF system is closely associated with the canceration of cells. Previous studies showed that the nuclear matrix in cancer cells had some distinctive irregular morphology which differed from those of normal cells obviously[15,16]. HMBA could inhibit the malignant proliferation of SMMC-7721 cells, change the expression activity of hepatocarcinoma cell-associated enzyme or antigen such as Gamma Glutamyl Transferase (γ-GT), α-fetoprotein (AFP), proliferating cell nuclear antigen (PCNA), and arrest the cells in G0/G1 phase. Furthermore, HMBA could up-regulate the expression of p21WAF1/CIP1 and p16 genes, down-regulate the activity of Cyclin D1-CDK4 and the transcription of c-myc gene which were necessary for cells entering into S phase, arrest the cells in G0/G1 phase, and induce the differentiation of SMMC-7721 cells[14,17]. This study further displayed that, after selective extraction, filaments of NM-IF system in SMMC-7721 cells were relatively few, not well-distributed and arranged irregularly, and the single filaments were few, the thick nuclear lamina was not closely associated with the nuclear matrix and intermediate filaments. It exhibited the rapid proliferation and the typical configuration characteristics of NM-IF system in malignant tumor cells. While in SMMC-7721 cells induced with HMBA, the nuclear matrix filaments and intermediate filaments interweaved into a regular and well-distributed meshwork in which the quantity of single filaments increased and differed in slender and thick form, moreover, the nuclear matrix filaments and intermediate filaments firmly fastened to the thin and compact fiber-like nuclear lamina. This morphology of NM-IF system was significantly different from those of rapidly proliferating SMMC-7721 cells but similar to those of some previously reported epithelial or normal cells[15,18], and was consistent with our previous studies in which we treated the human osteosarcoma cell line MG-63 and human gastric adenocarcinoma cell line MGC80-3 with retinotic acid (RA) respectively[12,19]. It showed that the morphology and function of NM-IF system in SMMC-7721 cells during its differentiation had undergone a reverse alteration. This alteration is not only important for the reversion of hepatocarcinoma cells, but also the indication of the differential expression of nuclear matrix proteins.
Through 2-D gel electrophoresis and analysis of MALDI-TOF-MS, we found 21 nuclear matrix proteins that were differentially expressed during the differentiation process. Among the changed spots, 8 were downregulated, one disappeared in the differentiated SMMC-7721 cells, whereas 11 spots were upregulated, and 1 emerged as a new protein spot that appeared in the differentiated cells. Nine of the 21 changed proteins were identified. Four proteins-mutant Pyst1, hypothetical protein, nucleophosmin1 and LBP were downregulated, whereas four proteins-eIF6, p44 subunit, β-tubulin and SIN3B were upregulated, and the SR2/ASF was only found in the differentiated SMMC-7721 cells. Previous researches showed that the composition of nuclear matrix in other cell lines varied apparently in the different stages of cell differentiation[20,21]. Our early studies also confirmed that the induced differentiation of human osteosarcoma cell line MG-63 and human gastric adenocarcinoma cell line MGC80-3 with HMBA was accompanied with changes of nuclear matrix proteins[13]. In this study, we proved for the first time that specific nuclear matrix proteins were accompanied with the differentiation of SMMC-7721 cells.
The identified differentially expressed proteins could be grouped into three classes: The first group was the important transcription factor that regulated the tumor cell proliferation associated gene expression, such as the basal transcription factor complex p44 subunit, the SIN3B protein which regulated the activation of Myc through interaction with Mad[22], the nucleophosmin which regulated gene transcription by binding with many factors of signal transduction[23-25], and the Pyst1 protein which played an important role in the MAPK signling[26]. The second was the splicing factor that reflected post-transcription level of the cell differentiation associated gene expression, such as the SFRS1 protein[27]. The third group was a kind of proteins that affected the synthesization, processing, and nuclear exportion of the ribosome, such as the multifuctional nucleophosmin, P27BBP/eIF6, and the LBP/P40[28,29]. Besides, one unknown protein (hypothetical protein) was found in this study and its function needs to be further illustrated. These identified nuclear matrix proteins were not only significant in the nuclear region, but also could influence the cell proliferation and differentiation through regulating the gene expression at transcriptional, mRNA processional or post-transcriptional level. Further characterization of those proteins and their interaction with the nuclear matrix network is important for revealing the mechanism of cancerization and understanding of the phenotypic reversion of tumor cells.
Previous studies showed that different stages of cell differentiation were characterized by different morphology of nuclear matrix. However, the relationship between alteration of morphology and function of nuclear matrix-intemediate filament system in cancer cells and its phenotypic reversion has not been well illustrated. The functions of important nuclear matrix proteins which influence the cancer cell differentiation have not been investigated in detail.
To identify differentially expressed nuclear matrix proteins and analyze their function in cancer cell differentiation is one of the most interesting hotspots in current studies of nuclear matrix.
The authors showed for the first time that the induced differentiation of human hepatocarcinoma cells had not only the morphological changes of nuclear matrix-intermediate filament system, but also the differential expressed nuclear matrix proteins which might be essential for cancer cell reversion.
Differentially expressed nuclear matrix proteins can be used as potential targets in both diagnosis and treatment of tumors.
Nuclear matrix: The residual proteinaseous structure that remains after the nuclei are depleted of the nuclear membranes, histones, soluble nuclear proteins and nucleic acids. The structures that remain in matrix preparations are the nuclear lamina, the residual nucleolus and the fibrillogranular network.
This is an interesting paper that links morphological and proteomical techniques in the study of nuclear matrix. The results are important for subsequent research on the function of those differentially expressed proteins.
S- Editor Liu Y L- Editor Ma JY E- Editor Lu W
| 1. | Tsutsui KM, Sano K, Tsutsui K. Dynamic view of the nuclear matrix. Acta Med Okayama. 2005;59:113-120. [PubMed] |
| 2. | Radichev I, Parashkevova A, Anachkova B. Initiation of DNA replication at a nuclear matrix-attached chromatin fraction. J Cell Physiol. 2005;203:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Iarovaia OV, Akopov SB, Nikolaev LG, Sverdlov ED, Razin SV. Induction of transcription within chromosomal DNA loops flanked by MAR elements causes an association of loop DNA with the nuclear matrix. Nucleic Acids Res. 2005;33:4157-4163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Girard-Reydet C, Grégoire D, Vassetzky Y, Méchali M. DNA replication initiates at domains overlapping with nuclear matrix attachment regions in the xenopus and mouse c-myc promoter. Gene. 2004;332:129-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Okorokov AL, Rubbi CP, Metcalfe S, Milner J. The interaction of p53 with the nuclear matrix is mediated by F-actin and modulated by DNA damage. Oncogene. 2002;21:356-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Gniadecki R, Olszewska H, Gajkowska B. Changes in the ultrastructure of cytoskeleton and nuclear matrix during HaCaT keratinocyte differentiation. Exp Dermatol. 2001;10:71-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Samaniego R, Jeong SY, de la Torre C, Meier I, Moreno Díaz de la Espina S. CK2 phosphorylation weakens 90 kDa MFP1 association to the nuclear matrix in Allium cepa. J Exp Bot. 2006;57:113-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A, Neale MH, Martin SJ, Latchman DS, Knight RA. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol Cell Biol. 2004;24:10593-10610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Taniura H, Kobayashi M, Yoshikawa K. Functional domains of necdin for protein-protein interaction, nuclear matrix targeting, and cell growth suppression. J Cell Biochem. 2005;94:804-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Ben-Yehoyada M, Ben-Dor I, Shaul Y. c-Abl tyrosine kinase selectively regulates p73 nuclear matrix association. J Biol Chem. 2003;278:34475-34482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Bosman FT. The nuclear matrix in pathology. Virchows Arch. 1999;435:391-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 12. | Li QF. Effect of retinoic acid on the changes of nuclear matrix in termediate filament system in gastric carcinoma cells. World J Gastroenterol. 1999;5:417-420. [PubMed] |
| 13. | Zhao CH, Li QF, Zhao Y, Niu JW, Li ZX, Chen JA. Changes of nuclear matrix proteins following the differentiation of human osteosarcoma MG-63 cells. Genomics Proteomics Bioinformatics. 2006;4:10-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Ouyang GL, Li QF, Peng XX, Hong SG. Differentiation of human hepatocarcinoma SMMC-7721 cells induced by HMBA. ShiYan ShengWu XueBao. 2001;34:269-273. [PubMed] |
| 15. | Fey EG, Wan KM, Penman S. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J Cell Biol. 1984;98:1973-1984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 408] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 16. | He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569-580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 342] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 17. | Ouyang GL, Li QF, Peng XX, Hong SG. Effects of HMBA on the expression of cell-cycle-associated genes in human hepatocarcinoma SMMC-7721 cells. ShiYan Shen Wu XueBao. 2002;35:173-178. [PubMed] |
| 18. | Fey EG, Penman S. Tumor promoters induce a specific morphological signature in the nuclear matrix-intermediate filament scaffold of Madin-Darby canine kidney (MDCK) cell colonies. Proc Natl Acad Sci USA. 1984;81:4409-4413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Zhao Y, Tang J, Zhao CH, Shi SL, Li QF. Observation of the effects of retinoic acid on the configurational changes of the NM-IF system and the alteration of nuclear matrix proteins in human osteosarcoma cell line MG-63. Xiamen Daxue Xuebao. 2006;45:6-10. |
| 20. | Alberti I, Barboro P, Barbesino M, Sanna P, Pisciotta L, Parodi S, Nicolò G, Boccardo F, Galli S, Patrone E. Changes in the expression of cytokeratins and nuclear matrix proteins are correlated with the level of differentiation in human prostate cancer. J Cell Biochem. 2000;79:471-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Jin ML, Zhang P, Ding MX, Yun JP, Chen PF, Chen YH, Chew YQ. Altered expression of nuclear matrix proteins in etoposide induced apoptosis in HL-60 cells. Cell Res. 2001;11:125-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | van Ingen H, Lasonder E, Jansen JF, Kaan AM, Spronk CA, Stunnenberg HG, Vuister GW. Extension of the binding motif of the Sin3 interacting domain of the Mad family proteins. Biochemistry. 2004;43:46-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 23. | Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 408] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 24. | Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 224] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 25. | Lambert B, Buckle M. Characterisation of the interface between nucleophosmin (NPM) and p53: potential role in p53 stabilisation. FEBS Lett. 2006;580:345-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Smith TG, Karlsson M, Lunn JS, Eblaghie MC, Keenan ID, Farrell ER, Tickle C, Storey KG, Keyse SM. Negative feedback predominates over cross-regulation to control ERK MAPK activity in response to FGF signalling in embryos. FEBS Lett. 2006;580:4242-4245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17:613-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 216] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 28. | Ceci M, Offenhäuser N, Marchisio PC, Biffo S. Formation of nuclear matrix filaments by p27(BBP)/eIF6. Biochem Biophys Res Commun. 2002;295:295-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 29. | Sato M, Kong CJ, Yoshida H, Nakamura T, Wada A, Shimoda C, Kaneda Y. Ribosomal proteins S0 and S21 are involved in the stability of 18S rRNA in fission yeast, Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2003;311:942-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |