Published online May 21, 2007. doi: 10.3748/wjg.v13.i19.2727
Revised: February 21, 2007
Accepted: March 8, 2007
Published online: May 21, 2007
AIM: To analyze the prevalence of germline MLH1 and MSH2 gene mutations and evaluate the clinical characteristics of Hungarian hereditary non-polyposis colorectal cancer (HNPCC) families.
METHODS: Thirty-six kindreds were tested for mutations using conformation sensitive gel electrophoreses, direct sequencing and also screening for genomic rearrangements applying multiplex ligation-dependent probe amplification (MLPA).
RESULTS: Eighteen germline mutations (50%) were identified, 9 in MLH1 and 9 in MSH2. Sixteen of these sequence alterations were considered pathogenic, the remaining two were non-conservative missense alterations occurring at highly conserved functional motifs. The majority of the definite pathogenic mutations (81%, 13/16) were found in families fulfilling the stringent Amsterdam I/II criteria, including three rearrangements revealed by MLPA (two in MSH2 and one in MLH1). However, in three out of sixteen HNPCC-suspected families (19%), a disease-causing alteration could be revealed. Furthermore, nine mutations described here are novel, and none of the sequence changes were found in more than one family.
CONCLUSION: Our study describes for the first time the prevalence and spectrum of germline mismatch repair gene mutations in Hungarian HNPCC and suspected-HNPCC families. The results presented here suggest that clinical selection criteria should be relaxed and detection of genomic rearrangements should be included in genetic screening in this population.
-
Citation: Papp J, Kovacs ME, Olah E. Germline
MLH1 andMSH2 mutational spectrum including frequent large genomic aberrations in Hungarian hereditary non-polyposis colorectal cancer families: Implications for genetic testing. World J Gastroenterol 2007; 13(19): 2727-2732 - URL: https://www.wjgnet.com/1007-9327/full/v13/i19/2727.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i19.2727
Hereditary non-polyposis colorectal cancer (HNPCC) is the most common form of hereditary colorectal cancer (CRC), accounting for 1%-5% of cases. This syndrome is characterized by early onset CRC and endometrial cancer, although the incidence of other malignant tumours, e.g. cancers of the stomach, urothelium, small bowel and ovarium, is also increased in HNPCC patients[1].
HNPCC is autosomal dominantly inherited and is associated with germline mutations in at least five mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS1 and PMS2)[2]. The identification of carriers is usually based on screening individuals from families fulfilling international criteria for the syndrome, namely Amsterdam criteriaIor II[3,4] or less stringent criteria referred to as the Bethesda guidelines[5].
Mutations in MLH1 (MIM# 120436) and MSH2 (MIM# 120435) are considered the major cause of HNPCC, since germline alterations of these genes have been found to be responsible for more than 90% of mutation carrier HNPCC families. According to the database of the International Society for Gastrointestinal Hereditary Tumors (InSiGHT; http://www.insight-group.org), currently more than 450 different pathogenic mutations have been described in these genes, accounting for approximately 750 HNPCC kindreds worldwide[6].
The majority of reported MLH1 and MSH2 mutations are nonsense, missense, or frameshift mutations as well as changes affecting splice sites. However, more recently, utilization of new techniques has led to the realization that in some populations a relatively large proportion of pathogenic alterations are genomic rearrangements, in most cases single or multi-exonic deletions or duplications inactivating MLH1 and/or MSH2[7,8]. The known mutations are scattered throughout the coding regions of these genes, although, in particular populations (e.g. in Finland, Poland and in the United States), some recurrent changes have been found and described as founder mutations[9-11].
Predictive genetic testing for germline mutations in MMR genes can allow identification of HNPCC families, and are thus of great importance for counselling, surveillance and management of at-risk patients. Nevertheless, since the reliability of the family history provided by patients is often incomplete and questionable, and because germline mutations are occasionally identified even in atypical HNPCC families[12], the requirements for establishing objective strategies of genetic testing are pressing. Therefore, assessing the prevalence and spectrum of MLH1 and MSH2 mutations in afflicted families in a given population is an essential first step in improving diagnosis of HNPCC, thus identifying individuals at high risk of colorectal and endometrial cancer.
From 1994 to 2004, clinical data and samples from 36 Hungarian HNPCC families were collected, of which 20 fulfilled the AmsterdamI-IIcriteria and 16 were classified as ‘pedigrees suggestive of HNPCC’. Although they did not meet internationally defined inclusion criteria, a suspicion of HNPCC was raised on the basis of their personal or family history being close to at least one of the modified Amsterdam criteria or the Bethesda guidelines[13]. The study was approved by the Institutional Ethical Board, and written informed consent was obtained from each patient following genetic counselling. In addition, 63 healthy subjects from the same population were analyzed for the presence of mutations with uncertain clinical significance.
Blood samples were obtained from all consenting subjects and DNA was extracted with the classic phenol-chloroform method. The entire MLH1 and MSH2 coding region and splice junctions were amplified by PCR from genomic DNA using primer sequences published earlier[14,15]. Systematic screening for point mutations in both genes was performed using conformation-sensitive gel electrophoreses (heteroduplex analysis and single-strand conformation polymorphism analysis) on all amplicons[16,17]. All samples showing altered migration patterns were subjected to direct bi-directional sequencing using an ABI 310 genetic analyzer (Applied Biosystems).
In addition to standard mutation analysis, both MLH1 and MSH2 coding regions were screened for genomic rearrangements using the multiplex ligation-dependent probe amplification (MLPA) technique[18]. In brief, 150-200 ng of genomic DNA in 5 μL Tris-EDTA was denatured, cooled, and incubated with the probe mix containing one probe for each MLH1/MSH2 exon and several control probes specific for sequences outside these genes. After probe hybridization, ligation using ligase-65 enzyme was done at 54°C for 15 min. Ligation products were then amplified by PCR using one fluorescently labelled primer. The PCR products were separated and analyzed with an ABI 310 genetic analyzer (Applied Biosystems). Peak heights for each fragment were compared to those of a control sample and deletions/duplications were suspected when peak intensities differed by more than 30%.
Since small sequence changes (SNPs, small insertions or deletions) close to the ligation site may also alter the peak size in MLPA, all exons showing copy number changes were directly sequenced. The presence of genomic rearrangements was confirmed by long PCR using the Expand Long Template PCR System (Roche) or Herculase Hotstart DNA polymerase (Stratagene) according to the manufacturers’ instructions. The PCR products were separated and sized by agarose gel electrophoresis and visualized by ethidium-bromide staining.
The mutation nomenclature used here complies with the recommendations of den Dunnen and Antonarakis[19,20]. Sequence variations were named in relation to the ATG codon in cDNA reference sequences (NM000249.2 and NM000251.1 for MLH1 and MSH2, respectively), while changes at the amino acid level were deduced from nucleotide alterations in relation to the protein reference sequences NP00240.1 for MLH1 and NP00242.1 for MSH2. All variants were ascertained by a replicate experiment.
Mutation analysis of affected members from 20 Hungarian HNPCC and 16 suspected HNPCC families revealed 18 germline alterations in 18 unrelated probands (50%), 9 in MLH1 and 9 in MSH2. Based on a literature review and checking the variants listed in the InSiGHT mutation database, we found that 9 out of 18 mutations have not been described previously. Sixteen of the sequence alterations were considered as pathogenic mutations either on the basis of published data or due to their predicted deleterious effects on the MSH2 or MLH1 protein such as insertions/deletions, nonsense changes, splice-site mutations and genomic rearrangements. The remaining two variants were missense alterations occurring at highly conserved motifs, one in MLH1 and the other in MSH2. Thirteen of the definite pathogenic mutations were found in members of HNPCC families fulfilling the stringent AmsterdamI/II criteria (including two deletions in MSH2 and one in MLH1). However, in three out of sixteen (19%) HNPCC-suspected families, a disease-causing alteration could be revealed, perhaps demonstrating the need for different inclusion decisions for this population. The MMR gene mutations, as well as the medical and family histories of patients, are shown in Table 1.
| ProbandID | Probandphenotype1 | Familyphenotype2 | Gene | Exon | Nucleotide position ofthe mutation | Effect/Consequence of themutation | |
| HFC003 | C25 | AI: | 9C, 1E | MLH1 | 2 | c.187G>A | p.D63N |
| HFC077 | C53 | AI: | 3C, 1S | MLH1 | 16 | c.1852_1854del | p.K618del |
| HFC100 | C39 | AI: | 5C, 3S, 1E | MLH1 | 18-19 | c.2078_2172del | p.E693_K724delAfsX8 |
| HFC075 | C33 | AI: | 3C | MSH2 | 1-2 | c.1-?_c.366+?del | no protein? |
| HFC048 | C31 | AI: | 6C | MSH2 | 3-7 | c.367-?_c.1226+?del | deletion of exons 3-7 |
| HFC066 | C22 | AI: | 6C, 1S | MSH2 | 4 | c.759_762del | p.M253_N254delIfsX20 |
| HFC108 | C30 | AI: | 5C, 1E | MSH2 | 7 | c.1226_1227del | p.Q409RfsX7 |
| HFC014 | C29 | AI: | 3C | MSH2 | 10 | c.1661+1G>T | splice defect? |
| HFC021 | C47 | AI: | 3C | MSH2 | 13 | c.2131C>T | p.R711X |
| HFC050 | C36 | AII: | 2C, 1I | MLH1 | 8 | c.677G>A | p.R226Q |
| HFC045 | C19 | AII: | 2C, 1E | MLH1 | 13 | c.1489_1490insC | p.R497fsX6 |
| HFE063 | E47 | AII: | 1C, 2E | MLH1 | 16 | c.1875T>G | p.Y625X |
| MPX050 | C36 | AII: | 2C, 2S, 1E, 1I | MLH1 | 19 | c.2141G>A | p.W714X |
| HFC017 | C27 | AII: | 3C, 3E | MSH2 | 12 | c.1861C>T | p.R621X |
| HFC072 | C35 | SH: | 1C | MLH1 | 13 | c.1411_1414del | p.K471_R472delDfsX19 |
| HFC104 | C39 | SH: | 2C | MLH1 | 16 | c.1852_1853delinsGC | p.K618A |
| HFC078 | C53 | SH: | 1C | MSH2 | 1 | c.146A>T | p.D49V |
| HFC074 | C45, 50, 55 | SH: | 4C | MSH2 | 15 | c.2620_2621ins115 | p.Y874CfsX3 |
Nine of the pathogenic changes identified in Hungarian families were found in the InSiGHT mutation database. Six of these were either nonsense mutations or small deletions, resulting in a premature stop codon predicted to be a subject of rapid mRNA degradation through nonsense-mediated decay (NMD)[21] or leading to the production of a truncated protein. One alteration (c.677G>A, p.R226Q), affecting the last nucleotide of MLH1 exon 8, presumably leads to splicing aberration[22]. This latter mutation was revealed in the proband of our family HFC050, diagnosed with colorectal carcinoma at age 36. Cosegregation with the disease phenotype could not be assessed.
The remaining two mutations were found in exon 16 of MLH1 (c.1852_1854del, p.K618del and c.1852_1853delinsGC, p.K618A), both affecting the same nonconserved lysine residue. Although K618A is one of the few changes listed as pathogenic and also as non-pathogenic in the InSiGHT database, previously published data indicate that both of these alterations are associated with an increased risk of colorectal cancer[23-25]. Furthermore, this C-terminal part of the MLH1 protein was shown to play a crucial role in protein-protein interactions, and a mutation of the same amino acid, K618T, caused nearly complete disruption of the interaction between MLH1 and MRE11, perhaps highlighting the potential function of this residue[26]. In our study, the non-conservative K618A variant was found in a small family (HFC104) showing two cases with early onset CRC (diagnosed at ages of 39 and 47 years) who were second degree relatives. DNA was available from only one of them, so cosegregation could not be evaluated. The in-frame deletion K618del was revealed in a family meeting the stringent Amsterdam I criteria (HFC077), but DNA sample was available only from the proband (diagnosed with CRC at age of 53).
None of these variants was present in 126 control chromosomes and also, no other possibly damaging sequence variant was found in any of these cases, further supporting the pathogenicity of these alterations.
Out of the nine novel sequence changes identified in Hungarian families, seven were considered pathogenic.
The 1661+1G>T substitution, found in a family fulfilling the Amsterdam I criteria (HFC014), could dramatically decrease the information content of the canonical 5’ splice site in intron 10 of MSH2 (ΔRi = -12.8), as determined by automated splicing mutation analysis (http://splice.cmh.edu)[27]. Although an RNA sample from the carrier proband was not available to test this hypothesis, the mutation is predicted to cause skipping of exon 10, thus leading to a frameshift and a premature stop codon.
In family HFE063, the proband was diagnosed with endometrial cancer at the age of 47. Mutation analysis revealed a substitution in exon 16 of MLH1. Sequencing showed this to be a c.1875T>G transversion, resulting in an immediate stop codon (p.Y625X). The mutation was also identified in the proband’s brother.
The proband of family HFC066 was diagnosed with cancer of the colorectum at the unusually early age of 22. His father died of CRC at the age of 32, and 5 other family members were affected with colorectal cancer (ages from 30 to 52 years). Conformation-sensitive mutation screening revealed a small deletion in exon 4 of MSH2. Sequence analysis identified the c.759_762delGAAT mutation, which results in frameshift and creates a stop 20 codons downstream. The mutation was also identified in the proband’s grandfather (CRC at the age of 52).
Family HFC074 was selected for mutation screening because, although the family phenotype did not fulfil Amsterdam criteriaI/II, the proband was diagnosed with three metachronous colorectal cancers (at the age of 45, 50 and 55 years) and his father was also affected (CRC at the age of 62). Testing for large genomic changes using MLPA analysis revealed a partial duplication of exon 15 of MSH2 following insertion of a guanine nucleotide. This aberration causes a frameshift, thus predicted to result in rapid degradation of RNA due to the nonsense-mediated decay surveillance mechanism.
MLPA screening of the proband (CRC diagnosed at the age of 33 years) from the AmsterdamIfamily HFC075 revealed a large genomic deletion affecting exons 1 and 2 of MSH2. The 5’ breakpoints of deletions are known to be very diverse[28,29]. In our case, the breakpoints for this deletion were not determined. However, since the deletion removes the transcription start site of the gene, it is predicted to result in a null allele.
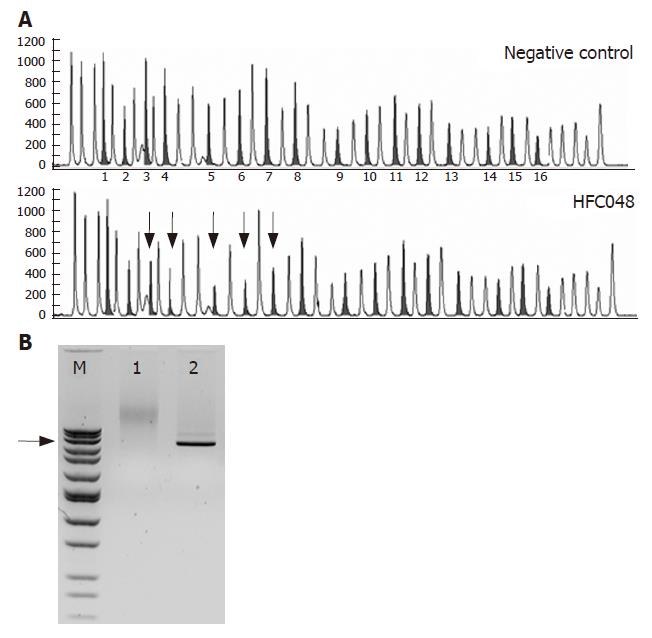
In the proband of our large AmsterdamIfamily HFC048, a large genomic deletion of MSH2 was detected using the MLPA method. Although the exact breakpoints have not been identified yet, long PCR analysis showed an approximately 30 kb deletion (Figure 1). This mutation is predicted to cause an out-of-frame deletion of exons 3-7, the resulting premature stop codon is presumably subjected to NMD.
Finally, MLPA analysis of family HFC100, a large kindred fulfilling AmsterdamIcriteria, showed a genomic deletion of MLH1, starting within exon 18 (codon 693) and ending in exon 19 (codon 724), thus removing parts of these exons and also the intervening intronic sequences. The result of this aberration is a deletion of 31 residues, and the appearance of a new frame terminating 8 residues downstream. Since this aberration removes the last exon-exon junction, nonsense-mediated decay cannot affect the stability of RNA, a slightly shorter protein is predicted to appear. The pathogenicity of this mutation is supported by the fact that (a) this most C-terminal part of MLH1 was shown to play a crucial role in protein-protein binding[26], and (b) in our family the mutation was also found in the proband’s mother affected by multiple primary cancers of the colon and stomach.
In family HFC003, the identified mutation is a single base substitution in exon 2 of the MLH1 gene. This mutation, c.187G>A, converts the aspartic acid residue at codon 63 to an asparagine (p.D63N) at the ATP binding pocket of the MLH1 protein. The substituted residue is highly conserved in a diverse class of ATPases, containing also type II topoisomerase, histidine kinases, gyrase B and Hsp90. In an earlier study, this p.D63N mutation was generated using site-directed mutagenesis and was shown to result not only in decreased activity but also in a dramatically reduced expression of the MutL-alpha complex containing the variant protein due to rapid degradation[30]. In our family, c.187G>A found in two CRC cases (father and son, diagnosed with CRC at the ages 40 and 25 years, respectively) was cosegregating with the disease, but was missing from seven other family members, who were asymptomatic at the age ranging from 19 to 57 years. These data, together with the findings mentioned above, strongly support the pathogenic nature of this mutation.
Additionally, in HFC078, a small family not fulfilling any of the accepted criteria (only one clinically confirmed colorectal cancer diagnosed at the age of 53 years), another substitution was revealed by mutation pre-screening and sequence analysis. The c.146A>T (p.D49V) transversion is situated in an N-terminal motif which is highly conserved in all MutS proteins throughout evolution. Mutations in this motif (GXFY(X)5DA) have been shown to play a crucial role in mismatch binding activity[31,32]. Unfortunately, since cosegregation with the disease could not be assessed in our atypical family, more data are needed to confirm or refute the pathogenicity of this variant.
Although none of these variants was found in 126 control chromosomes, these mutations are not to be used for predictive diagnostics, but considered unclassified variants until further functional tests prove or refute their causal association with the disease phenotype.
The current study, representing the first molecular characterization of HNPCC in Hungarian families, has expanded the known mutation spectrum of MSH2 and MLH1 genes. In summary, a total of 18 germline mutations were identified in 36 Hungarian cases (14 in HNPCC and 4 in suspected HNPCC families), half of them being novel (Table 2).
| Frequency of variant (%) | |||
| Pathogenic | Unclassified | Total | |
| HNPCC (AmsterdamI+ II) | 13/20 (65) | 1/20 (5) | 14/20 (70) |
| Suspected HNPCC | 3/16 (19) | 1/16 (6) | 4/16 (25) |
| Total | 16/36 (44) | 2/36 (6) | 18/36 (50) |
On the basis of the diffuse spectrum of point mutations and the relatively high frequency of genomic changes, we conclude that screening for rearrangements and mutations in the entire coding regions of these genes should be considered in this population. Moreover, since 25% of the suspected Hungarian HNPCC families were shown to carry a mutation in one of the MMR genes (three definitive pathogenic mutations and one mutation for which pathogenicity is assumed), we suggest that not only families fulfilling the stringent Amsterdam criteria, but also those following more relaxed guidelines should undergo genetic testing.
The authors are grateful to all the clinicians who referred patients and provided samples for this study. The authors thank Dr. Paul Brennan for critical reading of the manuscript and Gabi Varga, Marika Balogh and Judit Franko for their technical assistance.
Co-first-author: Janos Papp and Marietta E Kovacs
S- Editor Liu Y L- Editor Wang XL E- Editor Zhou T
| 1. | Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1426] [Cited by in RCA: 1370] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 2. | de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 429] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 3. | Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum. 1991;34:424-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1275] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 4. | Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1688] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 5. | Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 772] [Cited by in RCA: 712] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 6. | Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 331] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 7. | Nakagawa H, Hampel H, de la Chapelle A. Identification and characterization of genomic rearrangements of MSH2 and MLH1 in Lynch syndrome (HNPCC) by novel techniques. Hum Mutat. 2003;22:258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Taylor CF, Charlton RS, Burn J, Sheridan E, Taylor GR. Genomic deletions in MSH2 or MLH1 are a frequent cause of hereditary non-polyposis colorectal cancer: identification of novel and recurrent deletions by MLPA. Hum Mutat. 2003;22:428-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 9. | Nyström-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Järvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med. 1995;1:1203-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 206] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 10. | Kurzawski G, Suchy J, Kładny J, Safranow K, Jakubowska A, Elsakov P, Kucinskas V, Gardovski J, Irmejs A, Sibul H. Germline MSH2 and MLH1 mutational spectrum in HNPCC families from Poland and the Baltic States. J Med Genet. 2002;39:E65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, Nakagawa H, Geugien M, Jaghmohan-Changur S, Breukel C. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet. 2003;72:1088-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 142] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Beck NE, Tomlinson IP, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF. Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled. Br J Surg. 1997;84:233-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Umar A, Risinger JI, Hawk ET, Barrett JC. Testing guidelines for hereditary non-polyposis colorectal cancer. Nat Rev Cancer. 2004;4:153-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 114] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 14. | Wijnen J, Vasen H, Khan PM, Menko FH, van der Klift H, van Leeuwen C, van den Broek M, van Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer A. Seven new mutations in hMSH2, an HNPCC gene, identified by denaturing gradient-gel electrophoresis. Am J Hum Genet. 1995;56:1060-1066. [PubMed] |
| 15. | Beck NE, Tomlinson IP, Homfray T, Frayling I, Hodgson SV, Harocopos C, Bodmer WF. Use of SSCP analysis to identify germline mutations in HNPCC families fulfilling the Amsterdam criteria. Hum Genet. 1997;99:219-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Orban TI, Csokay B, Olah E. Sequence alterations can mask each other's presence during screening with SSCP or heteroduplex analysis: BRCA genes as examples. Biotechniques. 2000;29:94-98. [PubMed] |
| 17. | Ramus SJ, Kote-Jarai Z, Friedman LS, van der Looij M, Gayther SA, Csokay B, Ponder BA, Olah E. Analysis of BRCA1 and BRCA2 mutations in Hungarian families with breast or breast-ovarian cancer. Am J Hum Genet. 1997;60:1242-1246. [PubMed] |
| 18. | Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. [PubMed] |
| 19. | den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 20. | den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 644] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 21. | Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 866] [Cited by in RCA: 924] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 22. | Wijnen J, Khan PM, Vasen H, Menko F, van der Klift H, van den Broek M, van Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer EJ, Lindhout D. Majority of hMLH1 mutations responsible for hereditary nonpolyposis colorectal cancer cluster at the exonic region 15-16. Am J Hum Genet. 1996;58:300-307. [PubMed] |
| 23. | Farrington SM, Lin-Goerke J, Ling J, Wang Y, Burczak JD, Robbins DJ, Dunlop MG. Systematic analysis of hMSH2 and hMLH1 in young colon cancer patients and controls. Am J Hum Genet. 1998;63:749-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 103] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Liu T, Tannergård P, Hackman P, Rubio C, Kressner U, Lindmark G, Hellgren D, Lambert B, Lindblom A. Missense mutations in hMLH1 associated with colorectal cancer. Hum Genet. 1999;105:437-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Caldes T, Godino J, de la Hoya M, Garcia Carbonero I, Perez Segura P, Eng C, Benito M, Diaz-Rubio E. Prevalence of germline mutations of MLH1 and MSH2 in hereditary nonpolyposis colorectal cancer families from Spain. Int J Cancer. 2002;98:774-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Vo AT, Zhu F, Wu X, Yuan F, Gao Y, Gu L, Li GM, Lee TH, Her C. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 2005;6:438-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Nalla VK, Rogan PK. Automated splicing mutation analysis by information theory. Hum Mutat. 2005;25:334-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 28. | Di Fiore F, Charbonnier F, Martin C, Frerot S, Olschwang S, Wang Q, Boisson C, Buisine MP, Nilbert M, Lindblom A. Screening for genomic rearrangements of the MMR genes must be included in the routine diagnosis of HNPCC. J Med Genet. 2004;41:18-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Charbonnier F, Baert-Desurmont S, Liang P, Di Fiore F, Martin C, Frerot S, Olschwang S, Wang Q, Buisine MP, Gilbert B. The 5' region of the MSH2 gene involved in hereditary non-polyposis colorectal cancer contains a high density of recombinogenic sequences. Hum Mutat. 2005;26:255-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Räschle M, Dufner P, Marra G, Jiricny J. Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J Biol Chem. 2002;277:21810-21820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 92] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Malkov VA, Biswas I, Camerini-Otero RD, Hsieh P. Photocross-linking of the NH2-terminal region of Taq MutS protein to the major groove of a heteroduplex DNA. J Biol Chem. 1997;272:23811-23817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Dufner P, Marra G, Räschle M, Jiricny J. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSalpha is abolished by a single mutation in the hMSH6 subunit. J Biol Chem. 2000;275:36550-36555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 2.5] [Reference Citation Analysis (0)] |