Published online May 14, 2007. doi: 10.3748/wjg.v13.i18.2581
Revised: February 11, 2007
Accepted: March 26, 2007
Published online: May 14, 2007
AIM: To characterize and compare genotype profiles of H pylori strains isolated from patients with chronic gastritis and duodenal ulcer in western part of Turkey.
METHODS: A total of 46 patients [30 chronic gastritis (CG) and 16 duodenal ulcer (DU)] who had undergone endoscopy because of dyspeptic complaints were studied. The antral biopsy specimens were evaluated for the presence of H pylori by rapid urease test and culture, and the genotype profiles were determined by real-time PCR.
RESULTS: The cagA gene was observed in 43 (93.5%) isolates. The vacA s1m2 genotype was the predominant subtype, found in 63.3% and 68.7% of isolates in patients with CG and DU, respectively. Twenty (66.6%) isolates from patients with CG were iceA2 positive while the iceA1 was predominant in those with DU (68.8%). In terms of the association of the iceA alleles to other genes, both alleles were significantly associated with the cagA vacA s1m2 genotype.
CONCLUSION: The prevalent circulating genotypes in CG and DU were cagA vacA s1m2 iceA2 and cagA vacA s1m2 iceA1 genotype, respectively. It was found that cagA vacA s1m2 genotype seems to be common virulence factors in both CG and DU while iceA alleles show specificity for gastroduodenal pathologies in this study.
-
Citation: Caner V, Yilmaz M, Yonetci N, Zencir S, Karagenc N, Kaleli I, Bagci H.
H pylori iceA alleles are disease-specific virulence factors. World J Gastroenterol 2007; 13(18): 2581-2585 - URL: https://www.wjgnet.com/1007-9327/full/v13/i18/2581.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i18.2581
Bacterial colonization of the gastrointestinal tract of humans and other animals is still an interesting matter of study for microbiologists and gastroenterologists. H pylori is a fastidious Gram-negative bacterium that colonizes in human gastric mucosa. It had been estimated that more than half of the world's population is infected with this organism[1]. H pylori infection is a major cause of chronic gastritis (CG) and the small number of patients develop severe complications such as duodenal ulcer (DU), gastric ulcer, gastric cancer, and gastric non-Hodgkin's lymphoma of mucosa-associated lymphoid tissue (MALT lymphoma). Variation in clinical outcomes has been attributed to differences in environmental factors and host genetics, together with bacterial genotypes[2,3]. In this regard, the high genetic variability that characterizes H pylori and the influence of particular virulence genes (especially cagA, vacA, and iceA) on clinical outcome of H pylori infection have been reported from different geographic regions[4-8].
A strain-specific H pylori gene, cagA, was considered as a marker for the presence of a pathogenicity island (cag-PAI) which encodes several proteins implicated in the pathogenesis of H pylori[9]. Some of the genes in the island encode a type IV secretion system, which can translocate the CagA protein into target cells. It is also reported that the other genes are particularly associated with epithelial cell responses such as higher level of IL-8 and increased leukocyte infiltration[10,11]. It has been thought that cagA positive H pylori strains were associated with a more severe clinical outcome of H pylori infection[9] although there was no association between cagA status and the outcomes in Asia[6,12,13].
The vacA gene encodes the vacuolating cytotoxin which causes damage in epithelial cells. The gene possesses two regions; a signal region (s1 and s2 alleles) and a midregion (m1 and m2 alleles)[14,15]. Although the vacA gene is present in every H pylori strain only about half of the strains produce the active cytotoxin because the production of cytotoxin is related to the mosaic combination of s and m allelic types. The vacA s1m1 genotype is thought to be associated with more severe pathologies[16].
The other virulence gene, iceA which has a significant homology to a type II restriction endonuclease is also associated with H pylori infection[17]. Two allelic variants, iceA1 and iceA2, have been identified. The iceA2 positive strains have been reported more prevalant among patients with non-ulcer dyspepsia while H pylori strains possess iceA1 allele have been more prevalant in peptic ulcer disease[5,17].
The previous studies focused on determining the genotype profiles of H pylori isolates in different clinical outcomes of H pylori infection in Turkey were limited and carried out in different geographic regions which has different environmental factors, and nutritional habits, together with different lifestyle[8,18-20]. Therefore, the aims of this study were to characterize and to compare the genotype profiles of H pylori isolates in patients with chronic gastritis and duodenal ulcer in the western part of Turkey.
A total of 46 patients with dyspeptic complaints (30 women; mean age 49.5 and 16 men; mean age 46.9) who had undergone endoscopy in Pamukkale University Hospital were included. The exclusion criteria were briefly: treatment with antibiotics, non-steroidal anti-inflammatory drugs or proton pump inhibitors during the last 2 wk before endoscopy, having severe systematic disease, and uremic disease. The patients were divided into two groups as CG (n = 30) and DU (n = 16) according to the endoscopy reports. Written informed consent for participation was obtained from every patient before the study. The study protocol was approved in advance by the Human Institutional Review Board of Pamukkale University Medical School, and was performed in accor-dance with the Decleration of Helsinki. Antral biopsy specimes were evaluated for the presence of H pylori by rapid urease test and culture. The genotype profiles of H pylori isolates were determined by real-time PCR.
For rapid urease test, the specimens were inoculated into the CLOtest (Kimberly Clarck, USA). A positive result was recorded when the color changed from yellow to pink within 24 h.
For bacterial culture, the biopsy specimens were inocu-lated on Brain Heart Infusion Agar (Difco) containing 7% horse blood and H pylori selective supplement (Oxoid-SR 147E). The agar plates were incubated under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 37°C for up to 7 d. Colonies were identified as H pylori according to standard criteria including negative Gram's staining, typical cell morphology, and positive reactions for catalase, oxidase and urease.
H pylori genomic DNA was extracted from the biopsy specimens using the QIAamp DNA mini kit (Qiagen, Istanbul) as described by the manufacturer. One hundred microliters of elution buffer was used to resuspend the DNA. The genomic DNAs were stored at 4°C until used as a template in real-time PCR.
For real-time PCR, each reaction tube contained 2 μL of LightCycler FastStart Master SYBR Green I (Roche, Izmir), 12.4 μL of PCR-grade H2O, 1.6 μL of 25mmol/L MgCl2, 2 μL of a 10mmol/L concentration of primer set, and 2 μL of template DNA in a 20 μL PCR mixture. All oligonucleotide primers designed by Yamaoka et al[6] were used in real-time PCR and synthesized by TıbMolbiol (Berlin, Germany). The reaction protocol for cagA was as follows: an initial FastStart Taq DNA polymerase activation phase at 95°C for 10 min; a 35 cycle amplification phase consisting of a 95°C denaturation segment for 10 s, a 55°C annealing segment for 5 s, and a 72°C extension segment for 10 s. After completion of the amplification process, the reaction mixture was denaturated 95°C for 0 s, held at 65°C for 18 s, and then slowly heated to 95°C at a ramp rate of 0.2°C per second. The cagA real-time PCR protocol was used with little modifications for other oligonucleotide primers. At the end of the cycles, a cooling step at 40°C for 30 s was performed for each reaction.
All runs were included one negative DNA control consisting of PCR-grade water and two or more positive controls (HP 26695, HP J99 and some clinical isolates, a gift from Dr. Yamaoka, Baylor College, Texas, USA).
The χ2 test was used to compare differences in the prevalence of H pylori genotypes between groups. P values of < 0.05 were considered significant.
Patients were considered infected with H pylori infection if the biopsy specimens gave positive results in any one of the following tests: CLO test, culture, or real-time PCR. The rapid urease test and culture were positive in 78.2% and 86.9% of the specimens, respectively. Out of 46 specimens analyzed, all (100%) gave informative results by real-time PCR.
The cagA, vacA and iceA genotypes of H pylori isolates were determined by melting curve analysis of real-time PCR. The following isolates were also tested and found negative by real-time PCR: Campylobacter jejuni, Escherichia coli, Salmonella enterica serovar Enteritidis and Staphylococcus aureus (data was not shown). The melting temperatures for cagA, vacA s1, vacA s2, vacA m1, vacA m2, iceA1, and iceA2 were 80.08°C, 86.44°C, 76.15°C, 82.85°C, 84.59°C, 83.06°C, and 79.02°C, respectively.
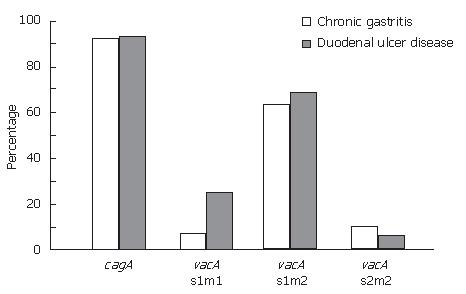
In this study, it was found that only three of 46 (6.5%) H pylori isolates were cagA negative, whereas the remaining 43 (93.5%) isolates were cagA positive. Of the cagA negative isolates, two were isolated from patients with CG. The prevalence of cagA gene was 93.3% and 93.75 in H pylori isolates in patients with CG and DU, respectively. There were no statistically significant differences between the cagA positivity and CG or DU (Figure 1).
All DNA extracts described in this study were positive for the vacA gene. For the vacA s and m region, 41/46 (89.1%) isolates were the type s1 and 38/46 (82.6%) isolates were the type m2. The vacA s1m2 genotype was the predominant subtype, being found in 63.3% of the isolates in patients with CG and 68.7% of those with DU. No vacA s2m1 genotype was determined in this study (Figure 1). No correlation between the vacA genotype and both gastroduodenal pathologies was observed. The presence of the vacA s1m2 genotype in combination with cagA were 73.3% and 68.7% of the isolates in CG and DU, respectively.
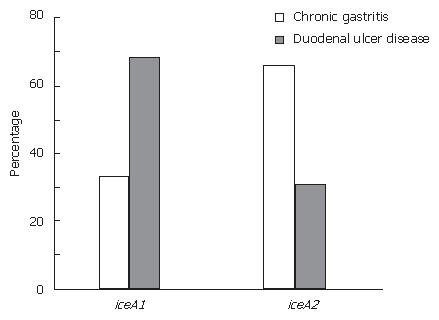
In this study, the iceA1 allele was detected in 45.7% (21/46) of the H pylori isolates, and the iceA2 allele was detected in 54.3% (25) of the isolates. The iceA1 allele was significantly associated with DU (68.8%, P < 0.05) while there was a significant relationship between iceA2 allele and CG (66.6%, P < 0.05) (Figure 2).
Several epidemiological studies have been reported the influence of particular virulence genes (especially cagA, vacA, and iceA) on clinical outcome of H pylori infection in different geographic regions[4-8]. This study was designed to characterize and to compare the genotype profiles of H pylori strains isolated from patients with chronic gastritis and duodenal ulcer in western part of Turkey.
In this study, the prevalence of cagA gene was 93.3% and 93.75 in H pylori isolates in patients with CG and DU, respectively. The result obtained from the isolates in the patients with DU is in agreement with other studies carried out in patients with DU in Turkey (85%-89%)[8,18]. However, the cagA prevalence in total is similar to those reported in Asia[6,12,21] and Ireland[22] but higher than those in Western countries[5,23,24]. Interestingly, the high prevalence of cagA (93.3%) in isolates of patients with CG was observed. The presence of cagA is known as a predictive marker for cag-PAI. Although intact cag-PAI was associated with the development of gastroduodenal pathology[25] it was also recently reported that this island was not intact in many strains across the world[26]. Therefore, the high prevalence of cagA in CG could not be explained precisely, a possible reason to explain this phenomenon is that the cagA positive strains with nonintact PAI might likely carry deleted or nonfunctional virulence genes[25].
The vacA gene was detected in all strains. The vacA s1m2 genotype predominant irrespective of the clinical outcome was also predominant in strains from Western countries including Turkey[4,18,22,27]. Aydin F et al[19] reported the vacA s1m1 genotype was the common vacA genotype in Black sea region located in Asian part of Turkey. The prevalence of vacA s1m1 genotype in our study was 6.6% and 25% of the strains in patients with CG and DU, respectively. The assesment of vacA gene mosaicism found only three out of the four possible combinations, the s2m1 mosaicism being never detected in our series, which was typically observed in strains from Western country including Turkey[18,19,22,24]. In this study, the cagA vacA s1m2 genotype was predominant genotype, and there was no significant relationship between both of the pathologies, indicating that the genotype was common virulence factors of H pylori.
Although the function of iceA gene is not clear, it is known that the expression of the gene is induced by contact between H pylori and the epithelial cells of the stomach[17]. The previous studies focused on the iceA genotyping in Turkey was limited. One of them reported that there was no significant association between the iceA genotype and clinical outcome of H pylori infection[20] while the other was found the iceA allele was significantly higher among patients with gastric cancer when compared to patients with non-ulcer dyspepsia[18]. In present study, most H pylori strains (66.6%) isolated from patients with chronic gastritis had iceA2 allele while iceA1 allele was predominant in those with duodenal ulcer disease (68.8%). This difference was statistically significant. In terms of the association of the iceA1 allele to DU, there is an agreement with the previous studies carried out in the USA[6], China[28], and Netherland[5] but in contrast to the results reported from Japan[6]. The high prevalence of the iceA2 allele in patients with CG was also observed in the USA[6]. Such differences the discrepant results between the iceA alleles and the clinical outcome of H pylori infection could be explained by the genetic heterogenity or to differences in the geographic locations as were previously reported for the other virulence genes[29,30].
In summary, the prevalent circulating genotypes in CG and DU in western part of Turkey were cagA vacA s1m2 iceA2 and cagA vacA s1m2 iceA1 genotype, respectively. It was also found that cagA vacAs1m2 genotype seems to be common virulence factors in both chronic gastritis and duodenal ulcer disease while iceA alleles show specificity for gastroduodenal ulcer disease. To understand the pathogenesis of the infection, the population genetics of H pylori together with host response including genetic predisposition and immune response, and environmental factors should also be considered.
The authors wish to acknowledge the financial support provided by a grant from the State Planning Organization in the Prime Ministry of Republic of Turkey (DPT-2003K120950). The authors also thank Dr. Nilay Sen Turk for statistical advace and Dario Papi for designing of all oligonucleotide primers. This work was presented in part at the XXXI.th Turkish Microbiology Congress, Kusadasi-Aydin, Turkey, 19-23 September 2004.
S- Editor Liu Y L- Editor Alpini GD E- Editor Liu Y
| 1. | Lambert JR, Lin SK, Aranda-Michel J. Helicobacter pylori. Scand J Gastroenterol Suppl. 1995;208:33-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 2. | Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;113:321-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 621] [Article Influence: 29.6] [Reference Citation Analysis (1)] |
| 3. | Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1317] [Cited by in RCA: 1343] [Article Influence: 58.4] [Reference Citation Analysis (1)] |
| 4. | Miehlke S, Yu J, Schuppler M, Frings C, Kirsch C, Negraszus N, Morgner A, Stolte M, Ehninger G, Bayerdörffer E. Helicobacter pylori vacA, iceA, and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease. Am J Gastroenterol. 2001;96:1008-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 412] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 6. | Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274-2279. [PubMed] |
| 7. | Li L, Graham DY, Gutierrez O, Kim JG, Genta RM, El-Zimaity HM, Go MF. Genomic fingerprinting and genotyping of Helicobacter pylori strains from patients with duodenal ulcer or gastric cancer from different geographic regions. Dig Dis Sci. 2002;47:2512-2518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Saribasak H, Salih BA, Yamaoka Y, Sander E. Analysis of Helicobacter pylori genotypes and correlation with clinical outcome in Turkey. J Clin Microbiol. 2004;42:1648-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 74] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93:14648-14653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1375] [Cited by in RCA: 1392] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 10. | Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 954] [Cited by in RCA: 963] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 11. | Zarrilli R, Ricci V, Romano M. Molecular response of gastric epithelial cells to Helicobacter pylori-induced cell damage. Cell Microbiol. 1999;1:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Pan ZJ, van der Hulst RW, Feller M, Xiao SD, Tytgat GN, Dankert J, van der Ende A. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol. 1997;35:1344-1347. [PubMed] |
| 13. | Maeda S, Ogura K, Yoshida H, Kanai F, Ikenoue T, Kato N, Shiratori Y, Omata M. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut. 1998;42:338-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 183] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 14. | Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570-10575. [PubMed] |
| 15. | Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1108] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 16. | Atherton JC, Peek RM, Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 405] [Article Influence: 14.5] [Reference Citation Analysis (1)] |
| 17. | Peek RM, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, Miller GG. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531-544. [PubMed] |
| 18. | Erzin Y, Koksal V, Altun S, Dobrucali A, Aslan M, Erdamar S, Dirican A, Kocazeybek B. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter. 2006;11:574-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Aydin F, Kaklikkaya N, Ozgur O, Cubukcu K, Kilic AO, Tosun I, Erturk M. Distribution of vacA alleles and cagA status of Helicobacter pylori in peptic ulcer disease and non-ulcer dyspepsia. Clin Microbiol Infect. 2004;10:1102-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Baglan PH, Bozdayi G, Ozkan M, Ahmed K, Bozdayi AM, Ozden A. Clarithromycin resistance prevalence and Icea gene status in Helicobacter Pylori clinical isolates in Turkish patients with duodenal ulcer and functional dyspepsia. J Microbiol. 2006;44:409-416. [PubMed] |
| 21. | Zhou W, Yamazaki S, Yamakawa A, Ohtani M, Ito Y, Keida Y, Higashi H, Hatakeyama M, Si J, Azuma T. The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS Immunol Med Microbiol. 2004;40:81-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Ryan KA, Moran AP, Hynes SO, Smith T, Hyde D, O'Morain CA, Maher M. Genotyping of cagA and vacA, Lewis antigen status, and analysis of the poly-(C) tract in the alpha(1,3)-fucosyltransferase gene of Irish Helicobacter pylori isolates. FEMS Immunol Med Microbiol. 2000;28:113-120. [PubMed] |
| 23. | Rudi J, Kolb C, Maiwald M, Kuck D, Sieg A, Galle PR, Stremmel W. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36:944-948. [PubMed] |
| 24. | Andreson H, Lõivukene K, Sillakivi T, Maaroos HI, Ustav M, Peetsalu A, Mikelsaar M. Association of cagA and vacA genotypes of Helicobacter pylori with gastric diseases in Estonia. J Clin Microbiol. 2002;40:298-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 25. | Nilsson C, Sillén A, Eriksson L, Strand ML, Enroth H, Normark S, Falk P, Engstrand L. Correlation between cag pathogenicity island composition and Helicobacter pylori-associated gastroduodenal disease. Infect Immun. 2003;71:6573-6581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Kauser F, Khan AA, Hussain MA, Carroll IM, Ahmad N, Tiwari S, Shouche Y, Das B, Alam M, Ali SM. The cag pathogenicity island of Helicobacter pylori is disrupted in the majority of patient isolates from different human populations. J Clin Microbiol. 2004;42:5302-5308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 72] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 27. | Van Doorn LJ, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz DM, Carneiro F, Vanderborght B, Pegado MD, Sanna R. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 299] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 28. | Lin HJ, Perng CL, Lo WC, Wu CW, Tseng GY, Li AF, Sun IC, Ou YH. Helicobacter pylori cagA, iceA and vacA genotypes in patients with gastric cancer in Taiwan. World J Gastroenterol. 2004;10:2493-2497. [PubMed] |
| 29. | Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 762] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 30. | Yamaoka Y, Orito E, Mizokami M, Gutierrez O, Saitou N, Kodama T, Osato MS, Kim JG, Ramirez FC, Mahachai V. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 2002;517:180-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 153] [Article Influence: 6.7] [Reference Citation Analysis (0)] |