Published online May 14, 2007. doi: 10.3748/wjg.v13.i18.2568
Revised: January 26, 2007
Accepted: February 8, 2007
Published online: May 14, 2007
AIM: To evaluate the methylation status of CDH1, FHIT, MTAP and PLAGL1 promoters and the association of these findings with clinico-pathological characteristics.
METHODS: Methylation-specific PCR (MSP) assay was performed in 13 nonneoplastic gastric adenocarcinoma, 30 intestinal-type gastric adenocarcinoma and 35 diffuse-type gastric adenocarcinoma samples from individuals in Northern Brazil. Statistical analyses were performed using the chi-square or Fisher's exact test to assess associations between methylation status and clinico-pathological characteristics.
RESULTS: Hypermethylation frequencies of CDH1, FHIT, MTAP and PLAGL1 promoter were 98.7%, 53.9%, 23.1% and 29.5%, respectively. Hypermethylation of three or four genes revealed a significant association with diffuse-type gastric cancer compared with nonneoplastic cancer. A higher hypermethylation frequency was significantly associated with H pylori infection in gastric cancers, especially with diffuse-type. Cancer samples without lymph node metastasis showed a higher FHIT hypermethylation frequency. MTAP hypermethylation was associated with H pylori in gastric cancer samples, as well as with diffuse-type compared with intestinal-type. In diffuse-type, MTAP hypermethylation was associated with female gender.
CONCLUSION: Our findings show differential gene methylation in tumoral tissue, which allows us to conclude that hypermethylation is associated with gastric carcinogenesis. MTAP promoter hypermethylation can be characterized as a marker of diffuse-type gastric cancer, especially in women and may help in diagnosis, prognosis and therapies. The H pylori infectious agent was present in 44.9% of the samples. This infection may be correlated with the carcinogenic process through the gene promoter hypermethylation, especially the MTAP promoter in diffuse-type. A higher H pylori infection in diffuse-type may be due to greater genetic predisposition.
-
Citation: Leal MF, Lima EM, Silva PNO, Assumpção PP, Calcagno DQ, Payão SLM, Burbano RR, Smith MAC. Promoter hypermethylation of
CDH1 ,FHIT ,MTAP andPLAGL1 in gastric adenocarcinoma in individuals from Northern Brazil. World J Gastroenterol 2007; 13(18): 2568-2574 - URL: https://www.wjgnet.com/1007-9327/full/v13/i18/2568.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i18.2568
Gastric cancer (GC) is the most frequent type of cancer and is the second most common cause of cancer death in the world[1]. In Northern Brazil, the State of Pará has a high incidence of this neoplasia and its capital, Belém, is ranked eleventh in terms of GC incidence rates among all cities in the world with cancer records[2]. Food may be related to the high incidence of this neoplasia in Pará, especially the high consumption of salt-conserved food, reduced use of refrigerators and little consumption of fresh fruit and vegetables[3].
The most successful and widely used classification of GC is that of Laurén[4], which is divided in two main types: diffuse and intestinal. Intestinal-type GC is usually called "epidemic", is more frequent, more related to environmental factors and associated with precancerous lesions, such as chronic gastritis, gastric atrophy, intestinal metaplasia and dysplasia. Diffuse-type have a poorer prognosis, are not associated with precancerous lesions and show invasive growth patterns[5].
Epigenetic events play a significant role in cancer development and progression. DNA methylation is the most studied epigenetic alteration, occurring through the addition of a methyl radical to the cytosine base adjacent to guanine. When DNA is methylated in the gene promoter region, genes are inactivated and silenced[6]. In cancer, epigenetic silencing leads to the aberrant silencing of normal tumor-suppressor functions.
Four genes were chosen for assessment of their functions and/or roles in gastric carcinogenesis: CDH1, FHIT, MTAP and PLAGL1.
E-cadherin, CDH1 product, is a homophilic cell adhesion protein. It has been proposed that loss of E-cadherin-mediated cell-cell adhesion is a prerequisite for tumor cell invasion and metastasis[7]. E-cadherin also has a possible role in modulating intracellular signaling, thus promoting tumor growth[8].
FHIT gene was identified in the most common fragile site, FRA3B[9]. FHIT is highly expressed in all normal epithelial tissues. FHIT is inactivated in about 60% of human tumors (ranging from 20% to 100%). Thus, FHIT is the most commonly altered gene in human cancer and in precancerous conditions[10]. Despite strong evidence of a FHIT tumor suppressor function, the specific signal pathways and mechanisms involved are still unknown.
MTAP is expressed ubiquitously in all normal cells[11]. Reduced MTAP expression may lead to activation of the polyamine biosynthetic enzyme ornithine decarboxylase (ODC)[12]. In gastric tissue, the ODC activity is elevated in premalignant lesions, which may be useful as biochemical markers of neoplastic proliferation[13]. Many malignant cells lack MTAP activity because of chromosomal loss or epigenetic regulation[14]. To our knowledge, the methylation status of MTAP has not been reported in GC.
PLAGL1 encodes a zinc-finger protein that regulate induction of apoptosis and G1 arrest[15] and contains transactivation and repressor activities[16]. Besides TP53, the identification of PLAGL1 provides the first example of a gene which combines concomitant induction of programmed cell death and cell arrest[17].
Identification of tumor specific epigenetic alterations can be used as a molecular marker of malignancy, which can lead to better diagnosis, prognosis and therapy. In this study, we evaluated the methylation status of CDH1, FHIT, MTAP and PLAGL1 promoters and hypermethylation frequency as well as their association with clinico-pathological characteristics.
The study included 78 samples of gastric tissue. Among them, 13 were nonneoplastic gastric mucosae, including 5 samples from GC patients (distant location of primary tumor) and 65 sporadic GC samples.
Eight normal mucosa samples (samples 1-8) were obtained from patients undergoing upper endoscopy to check for gastritis. All these samples were obtained from the antrum of stomach biopsies. The other nonneoplastic (samples 9-13) and GC samples were surgically obtained in Pará State João de Barros Barreto University Hospital (HUJBB). Patients had never been submitted to chemotherapy or radiotherapy prior to surgery, or had any other diagnosed cancer. All patients signed an informed consent with the approval of the ethics committee of HUJBB and of the Universidade Federal de São Paulo (UNIFESP). All 65 GC samples were classified according to Laurén[4]: 30 were intestinal (samples 14-43) and 35 were diffuse type (samples 44-78). Tumors were staged using standard criteria by TNM staging[18]. A rapid urease test was performed on all samples to detect H pylori.
Genomic DNA (2 μg) was modified by bisulfite treatment, converting unmethylated cytosines to uracils and leaving methylated cytosines unchanged. MSP was performed on treated DNA as previously described[19]. Specific primers for MSP (Table 1), located within CpG islands and previously described as associated with their genes expression[20-23], were designed with the assistance of Methprimer software[24].
| Gene | Sense | Antisense | Product size | TM |
| CDH1 | M-ATTCGAATTTAGTGGAATTAGAATC | M-CCCAAAACGAAACTAACGAC | 125 bp | 53°C |
| U-GGATTTGAATTTAGTGGAATTAGAATT | U-CTCCCCAAAACAAAACTAACAAC | 130 bp | ||
| FHIT | M-TTTTCGTTTTTGTTTTTAGATAAGC | M-AAAAATATACCCACTAAATAACCGC | 157 bp | 52°C |
| U-TGGTTTTTGTTTTTGTTTTTAGATAAGT | U-AAAATATACCCACTAAATAACCACC | 159 bp | ||
| MTAP | M-TGTTTTTTAGGAATTAAGGGAAATAC | M-AACTACAAAATCTAACCCGACGAC | 199 bp | 52°C |
| U-TTTTTAGGAATTAAGGGAAATATGT | U-CAACTACAAAATCTAACCCAACAAC | 197 bp | ||
| PLAGL1 | M-GTTTATTTTGGCGGAGATTTC | M-ACTAAACGACACCCACACGTC | 147 bp | 51°C |
| U-GGTTTATTTTGGTGGAGATTTTG | U-AAAAACTAAACAACACCCACACAT | 152 bp |
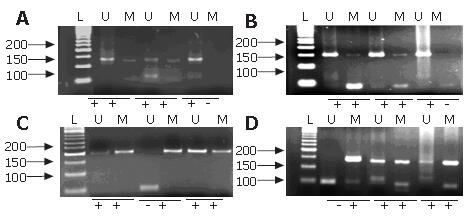
PCR reaction was carried out in a volume of 25 μL with 200 μmol/L of dNTPs, 200 μmol/L of MgCl2, 100 ng of DNA, 200 pmol/L of primers and 1 unit of AmpliTaq GOLD (Applied Biosystems, Foster City, CA). Initial denaturing was carried out for 3 min at 94°C, 35 cycles at 94°C for 30 s, at different temperatures with the primers for 45 s (Table 1) and 72°C for 30 s. This was followed by a final extension for 5 min at 72°C. PCR products were separated by 3% agarose gel containing 0.0004% ethidium bromide and visualized under UV illumination (Figure 1).
DNA from peripheral lymphocytes of two healthy individuals and water were used as negative controls. MSP results were scored when there was a clearly visible band on the electrophoresis gel with the methylated and unmethylatated primers[19]. Hypermethylation was considered present with methylated sequences for CDH1 and FHIT promoter or only methylated sequences for MTAP and PLAGL1 promoter.
Statistical analyses were performed using the χ2 test or Fisher's exact test to assess associations between the methylation status and frequency, and clinico-pathological characteristics. P values less than 0.05 were considered significant.
Of the 78 patients, 54 were male and 24 were female, and their mean age was 56 ± 12.31 years (range 20-76). According to Laurén's classification, 35 were diffuse and 30 were intestinal types. All GC samples were in advanced stage. Table 2 shows cases with their clinico-pathological characteristics. There was a significant association between diffuse-type and H pylori infection in our sample (P = 0.0142).
| Variable (n) | Samples | P | ||
| Nonneoplastic n (%) | Intestinal n (%) | Diffuse n (%) | ||
| Gender | ||||
| Male (54) | 9 (16.67) | 20 (37.03) | 25 (46.3) | 0.9176 |
| Female (24) | 4 (16.66) | 10 (41.67) | 10 (41.67) | |
| Age (yr) | ||||
| ≤ 50 (23) | 7 (30.43) | 7 (30.43) | 9 (39.14) | 0.1056 |
| > 50 (55) | 6 (10.91) | 23 (41.82) | 26 (47.27) | |
| Smoking status | ||||
| Smoker (27) | 1 (3.7) | 13 (48.15) | 13 (48.15) | 0.0717 |
| No smoker (51) | 12 (23.53) | 17 (33.33) | 22 (43.14) | |
| H pylori | ||||
| Present (36) | 6 (16.67) | 8 (22.22) | 22 (61.11) | 0.0142a |
| Absent (42) | 7 (16.67) | 22 (52.38) | 13 (30.95) | |
| Location | ||||
| Cardia (25) | - | 11 (44) | 14 (56) | 0.783 |
| Noncardia (40) | - | 19 (47.5) | 21 (52.5) | |
| T stage | ||||
| T1, T2 (15) | - | 7 (46.67) | 8 (53.33) | 0.9638 |
| T3, T4 (50) | - | 23 (46) | 27 (54) | |
| Lymph node metastasis | ||||
| Present (51) | - | 23 (45.1) | 28 (54.9) | 0.7445 |
| Absent (14) | - | 7 (50) | 7 (50) | |
| Distant metastasis | ||||
| Present (6) | - | 4 (66.67) | 2 (33.33) | 0.4649 |
| Absent (55) | - | 23 (41.82) | 32 (58.18) | |
| Unknown (4) | - | 3 (75%) | 1 (25%) | |
All samples analyzed showed at least one hypermethylated gene promoter. The hypermethylation frequency of CDH1, FHIT, MTAP and PLAGL1 promoter were 98.7%, 53.9%, 23.1% and 29.5%, respectively (Figure 2). Only one sample (sample 4), of a 20 years old patient, did not show CDH1 hypermethylation. The methylation number and frequency of CDH1, FHIT, MTAP and PLAGL1 in nonneoplastic and neoplastic samples are displayed in Table 3. Statistical analysis showed a tendency for increased PLAGL1 hypermethylation in GC samples (P = 0.0937) and in diffuse-type (P = 0.0807) compared with nonneoplastic samples.
| Samples | Methylation status | ||||||||
| CDH1 | FHIT | MTAP | PLAGL1 | ||||||
| M/U | U | M/U | U | M/U | M | P | M/U | M | |
| Normal tissue (13) | 12 (92.31) | 1 (7.69) | 8 (61.54) | 5 (38.46) | 11 (84.62) | 2 (15.38) | 12 (92.31) | 1 (7.69) | |
| GC (65) | 65 (100) | 0 (0) | 34 (52.31) | 31 (47.69) | 49 (75.38) | 16 (24.62) | 43 (66.15) | 22(33.85) | |
| Diffuse-type GC (35) | 35 (100) | 0 (0) | 17 (48.57) | 18 (56.25) | 22 (62.86) | 13 (37.14) | 0.0249a | 23 (65.71) | 12(34.29) |
| Intestinal-type GC (30) | 30 (100) | 0 (0) | 17 (56.67) | 13 (43.33) | 27 (90) | 3 (10) | 20 (66.6) | 10(33.33) | |
Table 4 shows the number of hypermethylated genes (1-2 or 3-4) in nonneoplastic tissue and GC samples. A tendency for hypermethylation of three or four genes in cancer samples compared to nonneoplastic samples was observed (P = 0.0959). Three or four hypermethylated genes were significantly more frequently detected in diffuse-type than in nonneoplastic mucosa (P = 0.0396). Neither of two peripheral lymphocyte samples showed hypermethylation (Figure 2).
| Samples (n) | Number of hypermethylated genes | P value | |
| 1 or 2 | 3 or 4 | ||
| Nonneplastic gastric tissue (13) | 12 (92.31%) | 1 (7.69%) | |
| Gastric cancer (65) | 44 (67.69%) | 21 (32.31%) | 0.0396b |
| Diffuse-type GC (35) | 21 (60%) | 14 (40%) | |
| Intestinal-type GC (30) | 23 (76.67%) | 7 (23.33%) | |
We analyzed whether DNA hypermethylation was associated with clinico-pathological characteristics, and detected a tendency for hypermethylation of three or four genes in tissue samples with H pylori infection (P = 0.0522). GC samples showed a significant association between higher hypermethylation frequency and H pylori infection (P = 0.0428). This association was also observed in diffuse-type samples (P = 0.0033). In diffuse-type, hypermethylation of three or four genes tended to be higher in female patients than in male (P = 0.0529) and in tumors located in the noncardia stomach region (P = 0.0805). MTAP hypermethylation was associated with H pylori infection in gastric samples (P = 0.0465). MTAP hypermethylation was also associated H pylori infection in GC samples (P = 0.0175), as well as with diffuse-type compared with intestinal-type (P = 0.0249). GC samples without lymph node metastasis were associated with FHIT hypermethylation (P = 0.0018). An association was observed between absence of lymph node metastasis and FHIT hypermethylation in diffuse-type GC (P = 0.0408), as well as between female patients and MTAP hypermethylation (P = 0.0310). Absence of lymph node metastasis was associated with FHIT hypermethylation in intestinal-type GC (P = 0.0104), as well as a tendency for PLAGL1 hypermethylation and larger tumor extension (T3 and T4) (P = 0.0637).
DNA hypermethylation of CpG islands was detected in all gastric samples, even in nonneoplastic mucosa, which can contain precursor cells for cancer and/or precancerous lesions[25].
The stomach is the organ that presents more methylated CpG island in nonneoplastic cells, along with age-related methylation that reflects increased CpG island methylation frequency in GC[26].
In our study, higher hypermethylation frequency was significantly associated with diffuse-type GC compared with nonneoplastic tissue. These findings confirmed those from Leung et al[27], which evaluated the methylation status of 5 genes in 28 samples of GC and their corresponding nonneoplasms.
We detected that 100% of GC and 92.3% of nonneo-plastic samples were hypermethylated in CDHI promoter (Table 3), frequencies higher than those in the literature. CDH1 methylation status was not significantly different between GC and nonneoplastic samples. This is the first study that has evaluated CDH1 methylation frequency in gastric tissue samples from a Brazilian population.
The highest CDH1 hypermethylation frequency described in the literature was 90% in advanced GC samples[28], as well as in nonneoplastic tissue[29]. However, Zazula et al[29] did not find significant differences between methylation patterns in 84 GC samples and their nonneo-plastic controls.
Waki et al[30] evaluated CDH1 methylation status in nonneoplastic gastric mucosa samples at autopsies. The authors did not see this gene methylation in nonneoplastic gastric cells from people who were 22 years and younger. However, CDH1 methylation was found in 86% of nonneoplastic gastric epithelia of people who were over 45 years old. Our findings showed only one nonneoplastic sample without CDH1 methylation from a 20 years old patient. Thus, our findings confirm those from Waki's study. The incidence of GC rises with age, pointing to an association between age-related methylation and GC development[30,31]. CDH1 hypermethylation in nonneoplastic gastric epithelia has also been frequently associated with H pylori infection[31-33].
In our sample, higher CDH1 hypermethylation frequency may result from the association of age, H pylori infection, advanced tumor and epidemiological factors, such as genetics constitution and diet.
In our study, FHIT hypermethylation frequency was 52.3% in GC samples (Table 2). This value is lower than those from two GC studies in the literature[34,35]. On the other hand, the FHIT hypermethylation frequency was 61.5% in nonneoplastic samples, which is higher than in other studies. Roa et al[34] reported that FHIT was methylated in 62% of 47 GC samples. Schildhaus et al[35] observed FHIT hypermethylation in all 6 advanced proximal GC and in 40% (2 of 5 samples) of normal gastric tissue, suggesting the FHIT hypermethylation is a precursor for GC.
In our study, FHIT promoter hypermethylation was detected in 80% (8/10) of nonneoplastic samples with gastritis or from GC patients, whereas none of the 3 nonneoplastic samples from subjects without gastritis and without GC were methylated (data not shown). Our findings suggest that FHIT hypermethylation may be associated with the start of gastric carcinogenesis, as well as in other organs[36].
Unmethylated sequences together with methylated sequences in CDH1 and FHIT promoter may be a result of allelic heterozygosity, clonal heterogeneity or contamination by inflammatory cells or stromal.
The methylation status of the MTAP promoter has never been analyzed in gastric tissue samples, while this promoter has been described in other neoplasias. However, methylation in nonneoplastic tissues has never been described[22,37-40]. Similarly, the MTAP promoter hypermethylation has never been analyzed by MSP. We detected methylated sequences for MTAP promoter in all samples, including nonneoplastic and lymphocytes samples. Thus, the difference found between MTAP methylation status in our work and those from the literature may be due to different CpG islands analyzed, as well as methodological differences.
We considered the MTAP promoter hypermethylation when only methylated sequences were observed, com-paring with the methylation status in peripheral blood lymphocytes. The MTAP promoter hypermethylation was in 24.6% of GC and 15.4% of nonneoplastic samples (Table 3).
In all samples methylated sequences of PLAGL1 was also observed, including nonneoplastic and lymphocyte samples. The literature shows only one study that evaluated PLAGL1 methylation status in gastric samples[23], where MSP was performed for PLAGL1 in 5 GC cell lines, 10 primary GC and one nonneoplastic sample. The authors observed methylation sequences in all neoplastic samples, while only one sample showed methylated and unmethylated sequences. The nonneoplastic sample presented only unmethylated sequences.
Our findings did not confirm those of Yamashita et al[23] for the nonneoplastic sample. However, PLAGL1 has been described as a maternal imprinted gene and hypermethylation leads to transcriptional silencing, as reported in other neoplasias[41]. Considered an imprinted gene, we found PLAGL1 hypermethylation in 33.9% of GC and 7.7% of nonneoplastic gastric samples (Table 3).
In our study, a higher hypermethylation frequency of CpG islands was found in H pylori infected samples, confirming findings from studies by El-Omar et al[42,43] and Hmadcha et al[44]. El-Omar et al[42,43] reported that interleukin-1 polymorphism led to an upregulation of interleukin-1 with H pylori infection and was associated with increased risk of GC. Furthermore, Hmadcha et al[44] described that interleukin-1 might induce gene methylation through the production of nitric oxide and the subsequent activation of DNA methyltransferase. It is possible that H pylori induces methylation through the production of interleukin-1 and hence the downstream activation of nitric oxide and DNA methyltransferase[31].
A higher hypermethylation frequency was associated with H pylori in diffuse-type, which can be related to its higher incidence in our samples of diffuse-type GC. H pylori gastritis is the only known precursor for diffuse-type GC[45].
In our study, a higher MTAP hypermethylation frequency was also associated with H pylori infection in gastric samples, especially in diffuse-type. MTAP hypermethylation in diffuse-type that is associated with H pylori infection may be due to a genetic predisposition. An association with female patients was also observed, which may be related to diffuse-type incidence, that is more frequent in female patients[46].
In our study, GC samples without lymph node metastasis were associated with FHIT hypermethylation. However, because of the small number of samples without lymph node metastasis in our study, further studies are necessary to confirm this association.
This is the first study that has evaluated the methy-lation status of CDH1, FHIT, MTAP and PLAGL1 promoters and their hypermethylation frequencies in gastric tissue samples in a population from Northern Brazil. The methylation status of MTAP promoter has never been investigated in gastric samples. Our findings show that hypermethylation is associated with gastric carcinogenesis. The H pylori infectious agent was present in 44.9% of samples and this infection may be a part of the carcinogenesis process through the gene promoter hypermethylation; especially the MTAP promoter in diffuse-type. Increased H pylori infection in diffuse-type may also be due to higher genetic predisposition.
S- Editor Liu Y L- Editor Roberts SE E- Editor Wang HF
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13286] [Cited by in RCA: 13558] [Article Influence: 677.9] [Reference Citation Analysis (1)] |
| 2. | Cancer Databases and Other Resources. International Agency for Research on Cancer (IARC) page. Available from: http: //www.iarc.fr. |
| 3. | Pereira LP, Waisberg J, André EA, Zanoto A, Mendes Júnior JP, Soares HP. Detection of Helicobacter pylori in gastric cancer. Arq Gastroenterol. 2001;38:240-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. an Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] |
| 5. | Huntsman DG, Carneiro F, Lewis FR, MacLeod PM, Hayashi A, Monaghan KG, Maung R, Seruca R, Jackson CE, Caldas C. Early gastric cancer in young, asymptomatic carriers of germ-line E-cadherin mutations. N Engl J Med. 2001;344:1904-1909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 270] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 6. | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5074] [Cited by in RCA: 4880] [Article Influence: 212.2] [Reference Citation Analysis (0)] |
| 7. | Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198:11-26. [PubMed] |
| 8. | Chan AO. E-cadherin in gastric cancer. World J Gastroenterol. 2006;12:199-203. [PubMed] |
| 9. | Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3; 8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 736] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 10. | Pekarsky Y, Palamarchuk A, Huebner K, Croce CM. FHIT as tumor suppressor: mechanisms and therapeutic opportunities. Cancer Biol Ther. 2002;1:232-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Hori H, Tran P, Carrera CJ, Hori Y, Rosenbach MD, Carson DA, Nobori T. Methylthioadenosine phosphorylase cDNA transfection alters sensitivity to depletion of purine and methionine in A549 lung cancer cells. Cancer Res. 1996;56:5653-5658. [PubMed] |
| 12. | Subhi AL, Diegelman P, Porter CW, Tang B, Lu ZJ, Markham GD, Kruger WD. Methylthioadenosine phosphorylase regulates ornithine decarboxylase by production of downstream metabolites. J Biol Chem. 2003;278:49868-49873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Okuzumi J, Yamane T, Kitao Y, Tokiwa K, Yamaguchi T, Fujita Y, Nishino H, Iwashima A, Takahashi T. Increased mucosal ornithine decarboxylase activity in human gastric cancer. Cancer Res. 1991;51:1448-1451. [PubMed] |
| 14. | Wild PJ, Meyer S, Bataille F, Woenckhaus M, Ameres M, Vogt T, Landthaler M, Pauer A, Klinkhammer-Schalke M, Hofstaedter F. Tissue microarray analysis of methylthioadenosine phosphorylase protein expression in melanocytic skin tumors. Arch Dermatol. 2006;142:471-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 15. | Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814-2825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 185] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 16. | Hoffmann A, Ciani E, Boeckardt J, Holsboer F, Journot L, Spengler D. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol Cell Biol. 2003;23:988-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Kas K, Voz ML, Hensen K, Meyen E, Van de Ven WJ. Transcriptional activation capacity of the novel PLAG family of zinc finger proteins. J Biol Chem. 1998;273:23026-23032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 121] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 18. | Sobin LH, Wittekind CH. TNM classification of malignant tumours. 6th ed. New York: Wiley-Liss 2002; . |
| 19. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4249] [Article Influence: 146.5] [Reference Citation Analysis (0)] |
| 20. | Oue N, Mitani Y, Motoshita J, Matsumura S, Yoshida K, Kuniyasu H, Nakayama H, Yasui W. Accumulation of DNA methylation is associated with tumor stage in gastric cancer. Cancer. 2006;106:1250-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 115] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 21. | Tanaka H, Shimada Y, Harada H, Shinoda M, Hatooka S, Imamura M, Ishizaki K. Methylation of the 5' CpG island of the FHIT gene is closely associated with transcriptional inactivation in esophageal squamous cell carcinomas. Cancer Res. 1998;58:3429-3434. [PubMed] |
| 22. | Behrmann I, Wallner S, Komyod W, Heinrich PC, Schuierer M, Buettner R, Bosserhoff AK. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am J Pathol. 2003;163:683-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2'-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 201] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 24. | Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1881] [Cited by in RCA: 2093] [Article Influence: 91.0] [Reference Citation Analysis (0)] |
| 25. | Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, Kitano S, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 224] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 26. | Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427-5440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 862] [Cited by in RCA: 886] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 27. | Leung WK, Yu J, Ng EK, To KF, Ma PK, Lee TL, Go MY, Chung SC, Sung JJ. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91:2294-2301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 28. | Zheng ZH, Sun XJ, Ma MC, Hao DM, Liu YH, Sun KL. Studies of promoter methylation status and protein expression of E-cadherin gene in associated progression stages of gastric cancer. YiChuan XueBao. 2003;30:103-108. [PubMed] |
| 29. | Zazula M, Ferreira AM, Czopek JP, Kolodziejczyk P, Sinczak-Kuta A, Klimkowska A, Wojcik P, Okon K, Bialas M, Kulig J. CDH1 gene promoter hypermethylation in gastric cancer: relationship to Goseki grading, microsatellite instability status, and EBV invasion. Diagn Mol Pathol. 2006;15:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 31. | Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, Hui WM, Rashid A, Kwong YL. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 221] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 32. | Liu YC, Shen CY, Wu HS, Chan DC, Chen CJ, Yu JC, Yu CP, Harn HJ, Shyu RY, Shih YL. Helicobacter pylori infection in relation to E-cadherin gene promoter polymorphism and hypermethylation in sporadic gastric carcinomas. World J Gastroenterol. 2005;11:5174-5179. [PubMed] |
| 33. | Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, Cheng VY, Ng EK, Sung JJ. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216-3221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 34. | Roa JC, Anabalón L, Roa I, Tapia O, Melo A, Villaseca M, Araya JC. Promoter methylation profile in gastric cancer. Rev Med Chil. 2005;133:874-880. [PubMed] |
| 35. | Schildhaus HU, Kröckel I, Lippert H, Malfertheiner P, Roessner A, Schneider-Stock R. Promoter hypermethylation of p16INK4a, E-cadherin, O6-MGMT, DAPK and FHIT in adenocarcinomas of the esophagus, esophagogastric junction and proximal stomach. Int J Oncol. 2005;26:1493-1500. [PubMed] |
| 36. | Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti MA. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1998;58:5032-5037. [PubMed] |
| 37. | Berasain C, Hevia H, Fernández-Irigoyen J, Larrea E, Caballería J, Mato JM, Prieto J, Corrales FJ, García-Trevijano ER, Avila MA. Methylthioadenosine phosphorylase gene expression is impaired in human liver cirrhosis and hepatocarcinoma. Biochim Biophys Acta. 2004;1690:276-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Furuta J, Umebayashi Y, Miyamoto K, Kikuchi K, Otsuka F, Sugimura T, Ushijima T. Promoter methylation profiling of 30 genes in human malignant melanoma. Cancer Sci. 2004;95:962-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 39. | Hellerbrand C, Mühlbauer M, Wallner S, Schuierer M, Behrmann I, Bataille F, Weiss T, Schölmerich J, Bosserhoff AK. Promoter-hypermethylation is causing functional relevant downregulation of methylthioadenosine phosphorylase (MTAP) expression in hepatocellular carcinoma. Carcinogenesis. 2006;27:64-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Wang Y, Yu Q, Cho AH, Rondeau G, Welsh J, Adamson E, Mercola D, McClelland M. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia. 2005;7:748-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 82] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 41. | Abdollahi A. LOT1 (ZAC1/PLAGL1) and its family members: mechanisms and functions. J Cell Physiol. 2007;210:16-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 42. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1676] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 43. | El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, Stanford JL, Mayne ST, Goedert J, Blot WJ. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193-1201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 672] [Cited by in RCA: 676] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 44. | Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J Exp Med. 1999;190:1595-1604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 165] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Vauhkonen M, Vauhkonen H, Sipponen P. Pathology and molecular biology of gastric cancer. Best Pract Res Clin Gastroenterol. 2006;20:651-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 115] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 46. | Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003;3:592-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 249] [Article Influence: 11.3] [Reference Citation Analysis (0)] |