Published online May 7, 2007. doi: 10.3748/wjg.v13.i17.2442
Revised: February 2, 2007
Accepted: March 12, 2007
Published online: May 7, 2007
Since the discovery of HCV in 1989, the lack of a cell culture system has hampered research progress on this important human pathogen. No robust system has been obtained by empiric approaches, and HCV cell culture remained hypothetical until 2005. The construction of functional molecular clones has served as a starting point to reconstitute a consensus infectious cDNA that was able to transcribe infectious HCV RNAs as shown by intrahepatic inoculation in a chimpanzee. Other consensus clones have been selected and established in a human hepatoma cell line as replicons, i.e. self-replicating subgenomic or genomic viral RNAs. However, these replicons did not support production of infectious virus. Interestingly, some full-length replicons could be established without adaptive mutations and one of them was able to replicate at very high levels and to release virus particles that are infectious in cell culture and in vivo. This new cell culture system represents a major breakthrough in the HCV field and should enable a broad range of basic and applied studies to be achieved.
- Citation: Duverlie G, Wychowski C. Cell culture systems for the hepatitis C virus. World J Gastroenterol 2007; 13(17): 2442-2445
- URL: https://www.wjgnet.com/1007-9327/full/v13/i17/2442.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i17.2442
Since the discovery of the hepatitis C virus (HCV) via molecular cloning in 1989[1], its propagation in cell culture has been a major goal for virologists worldwide. All human hepatitis viruses are very difficult to grow in cell culture, and indeed no such system is currently available for the hepatitis B virus (HBV). This problem is also linked to the nature of the host cell, since highly differentiated human hepatocytes are very difficult to maintain in cell culture. However, it has been demonstrated that blood or serum from chronically infected HCV patients is infectious in vivo and does contain infectious virions suitable for the in vitro infection of cultured hepatocytes.
In 1992, Shimizu et al[2] were the first to report successful HCV infection of the human T-lymphocyte MOLT-4Ma and HPB-Ma lines which had been pre-infected with murine retroviruses. In 1995, Kato et al[3] reported similar results with the MT-2 cell line pre-infected with HTLV-I. Use of clone 10-2 of the HPB-Ma cell line enabled a one-year follow-up of the infection, with neutralization by antibodies, inhibition of replication by interferon, visualisation of 50 nm particles by immuno-electronmicroscopy and analysis of the virion density[4]. However, in these and other cell lines (Daudi, PBMC, etc.), viral replication could be only intermittently detected by RT-PCR. The human hepatoma cell lines Huh-7 and Hep-G2 could support HCV replication, as did other hepatocytic cell lines immortalized by the SV-40 T antigen[5]. Ito et al[6] have cultured primary hepatocytes from chronically infected HCV patients and have obtained relatively high viral titers in both the cells and the supernatant medium. Primary chimpanzee and human hepatocytes are permissive to HCV for the very limited time during which these cells are usable[7-9]. These data indicate that HCV replication (at least) was possible in hepatocyte and lymphocyte cell lines. However, these systems were not reliable and could not be used as models for studying the viral cycle in detail or for screening antiviral drugs. These goals should be achieved by using viral and cellular clones under stable infection conditions[10]. Reverse genetics studies are also very helpful, and it is noteworthy that viral genomes are generally small enough to be engineered by today's oligonucleotide ligation techniques, marking the passage from macromolecular chemistry to "life"[11].
HCV is a member of the Flaviviridae and harbours an envelope with two glycoproteins and a nucleocapsid containing a positive single-strand RNA genome of about 9600 nucleotides. The positive viral RNA (vRNA) has the same polarity as mRNA and can be directly translated in order to express the full set of viral proteins and initiate the viral life cycle. The vRNAs are generated by transcription of cloned, complementary DNA (cDNA) obtained by reverse transcription. The first cloned HCV genomes were incomplete, and a novel sequence at the 3' terminus of the hepatitis C virus was only discovered in 1995 by use of oligonucleotide ligation or synthesis at the 3'end of the HCV genome[12,13].
The first "infectious" cDNA from HCV genotype 1 was successfully obtained by two different groups in 1997[14,15]. The HCV genome (called "H77") was cloned from the serum of an infected patient with a high viral titer. The sequences of many different clones were aligned, a master consensus sequence was established and a full-length HCV consensus sequence was then generated. It was supposed that the viral quasispecies contained a majority of functional genomes with a minority of defective ones, as observed for other viruses[16]. Thus, the establishment of a master consensus sequence is one approach to detecting deleterious mutations in defective virions or mutations introduced by amplification and/or molecular cloning. The identification of an infectious cDNA was successfully achieved, since the transcribed RNAs were able to infect the chimpanzee when inoculated intrahepatically. Other full-length genomic HCV cDNAs have since been established but none of these clones have been adapted to cell culture, as only very low levels of replication were reported[15,17,18].
New strategies based on the subgenomic selectable HCV replicon were developed in order to enable the selection of cell clones containing autonomously replicating HCV RNAs. In 1999, Lohmann et al[19] established the first HCV genotype 1b replicon in the Huh-7 hepatoma cell line. Replicons are subgenomic constructs expressing the viral replicase complex and capable of autonomous viral replication. As with other positive RNA viral genomes, the authors constructed bicistronic vectors-allowing the exact transcription of the HCV 5' and 3' non-translated regions (NTRs)-and then used (1) the HCV internal ribosomal entry site (IRES) to ensure translation of the neomycin phosphotransferase gene (neo) and (2) the EMCV IRES to ensure translation of the HCV replicase complex (at least from NS3 to NS5B). Thus, neomycin-resistant cell clones containing replicating HCV RNAs were then selected. This successful approach enabled the selection of highly permissive cell clones (such as Huh-7.5 and Huh-7-Lunet[20,21]) and adaptive viral mutations (involving principally the NS3, NS4B and NS5A regions[22]). Several of these mutations alter the phosphorylation state of NS5A, the hyperphosphorylated form of which appears to be deleterious for efficient HCV replication[23,24]. These replicons have been very valuable tools for studying HCV replication and for testing antiviral drugs against HCV's major target enzymes, i.e. NS3 helicase, NS3/4A protease and NS5B polymerase. Full-length genomic replicons using the consensus approach (such as the wild type Con-1) failed to produce infectious particles when tested, since very low numbers of viral particles were obtained[25]. Indeed, the different markers enabled the selection of replicative genomic sequences but were not appropriate for selecting for viral particle secretion[26]. Selective pressure acts on the replicase complex (involving most of HCV's non-structural proteins) but not on structural proteins. Furthermore, at least some adaptive mutations seemed to optimize viral replication and impair viral particle production. Interestingly, some replicons were reported as being free of adaptive mutations and were able to replicate at a very high rate[27].
Eventually, one of the above-mentioned full-length replicons was found to release viral particles[28]. The replicon was a HCV genotype 2a clone called JFH-1, constructed by Takaji Wakita's group and isolated from a Japanese patient with fulminant hepatitis[29]. It is the first authentic HCV clone considered by the scientific community as capable of growing in cell culture. Transfection of the full-length JFH-1 genome into Huh-7 cells leads to the production of HCV particles that are infectious for naive cells and for chimpanzees. Given that full-length genomic RNA derived from other clones of the same genotype or different HCV genotypes was again not infectious in a cell culture system, chimeric viruses were constructed successfully. Hence, the sequences coding for the HCV JFH-1 structural proteins were replaced by the corresponding sequences from different HCV genotypes[30,31]. The HCV chimera's junction was situated in the NS2 protein, just after the first transmembrane domain in a junction region which has also been identified in the rare, naturally-occurring recombinant forms of HCV (generally found in isolates from intravenous drug users[32]). However, the viral titer obtained for most constructs was low, with the exception of the FL-J6-JFH1 chimeric 2a-2a recombinant virus. This homologous, chimeric form was shown to be infectious in the chimpanzee, and the virus recovered from the infected animal was infectious in cell culture. Comparison of the viral titers obtained by Dr. Wakita's group with those observed by other groups indicates that cell culture conditions may influence viral production. Continuous passages of infected cells or successive infections of naive cells with the supernatant of infected cells leads to an increase in viral production. This finding suggests that certain mutations are probably selected during the infection. Determinants present in the structural proteins may be important for viral release and infection in cell culture. Thus, a detailed study of the key features enabling operation of the complete HCV cell cycle should enable researchers to cultivate other isolates. Very recently, a genotype 1a (H77) virus production system producing low viral titers has been described[33].
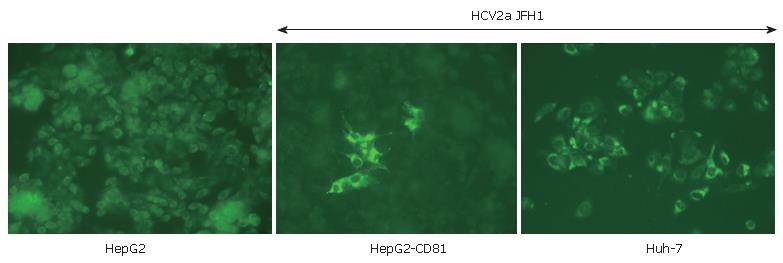
Another improvement concerns the cell line used for HCV culturing. Certain clones of the Huh-7 cell line were found to far better than others for growing HCV. "HCV-cured clones" produced by antiviral agents and Huh-7 subclones (such as Huh-7.5 and now Huh-7.5.1[34] or Huh-7-Lunet) support more efficient viral replication and production. Detailed studies have shown that some important features (such as high levels of CD81 receptor expression or perhaps SR-B1 or claudin-1 very recently identified by the Rice's group) could increase viral pro-duction and spreading. Indeed, as shown in Figure 1, HepG2 cells which are not usually susceptible to HCV can be infected by the JFH1 virus when they express CD81[30]. Furthermore, defects in innate immunity (such as deficiencies in interferon induction or production) could increase the cell line's permissivity[35,36]. Identification of new cell lines of hepatic (or perhaps lymphocyte) origin could improve our understanding of certain phy-siopathological aspects of hepatitis C. Equally, HCV pro-duction in cell lines from other species (such as murine cell lines) could help to establish a small animal model of HCV infection.
The new cell culture system represents a major breakthrough in the HCV field and should enable a broad range of fundamental and applied studies to be achieved, including the production of classic HCV vaccines for efficacy testing. Detailed cellular, molecular and reverse genetics studies are also needed to complete our knowledge of all aspects of HCV's biology.
The identity of the infectious form of HCV in "real life" remains open to debate because HCV in sera is strongly associated with lipoproteins; certain features (such as the infectious form's density) appear to differ from those seen in the HCV particles produced in cell culture systems (HCVcc). New information concerning the receptor(s) involved and other important determinants of permissivity will be valuable for improving the culture system. These new insights could enable researchers to grow clinical HCV isolates and study their particular features.
S- Editor Wang J E- Editor Ma WH
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4657] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 2. | Shimizu YK, Iwamoto A, Hijikata M, Purcell RH, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477-5481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 219] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 3. | Kato N, Nakazawa T, Mizutani T, Shimotohno K. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem Biophys Res Commun. 1995;206:863-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 84] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Shimizu YK, Purcell RH, Yoshikura H. Correlation between the infectivity of hepatitis C virus in vivo and its infectivity in vitro. Proc Natl Acad Sci USA. 1993;90:6037-6041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 116] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 5. | Seipp S, Mueller HM, Pfaff E, Stremmel W, Theilmann L, Goeser T. Establishment of persistent hepatitis C virus infection and replication in vitro. J Gen Virol. 1997;78:2467-2476. [PubMed] |
| 6. | Ito T, Mukaigawa J, Zuo J, Hirabayashi Y, Mitamura K, Yasui K. Cultivation of hepatitis C virus in primary hepatocyte culture from patients with chronic hepatitis C results in release of high titre infectious virus. J Gen Virol. 1996;77:1043-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 226] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 8. | Iacovacci S, Manzin A, Barca S, Sargiacomo M, Serafino A, Valli MB, Macioce G, Hassan HJ, Ponzetto A, Clementi M. Molecular characterization and dynamics of hepatitis C virus replication in human fetal hepatocytes infected in vitro. Hepatology. 1997;26:1328-1337. [PubMed] |
| 9. | Fournier C, Sureau C, Coste J, Ducos J, Pageaux G, Larrey D, Domergue J, Maurel P. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J Gen Virol. 1998;79:2367-2374. [PubMed] |
| 10. | Bartenschlager R. Hepatitis C virus molecular clones: from cDNA to infectious virus particles in cell culture. Curr Opin Microbiol. 2006;9:416-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 512] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 12. | Tanaka T, Kato N, Cho MJ, Shimotohno K. A novel sequence found at the 3' terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 217] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 13. | Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3' terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363-3371. [PubMed] |
| 14. | Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 549] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 15. | Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738-8743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 420] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 16. | Moormann RJ, van Gennip HG, Miedema GK, Hulst MM, van Rijn PA. Infectious RNA transcribed from an engineered full-length cDNA template of the genome of a pestivirus. J Virol. 1996;70:763-770. [PubMed] |
| 17. | Beard MR, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar DV, Lemon SM. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 116] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 18. | Lanford RE, Lee H, Chavez D, Guerra B, Brasky KM. Infectious cDNA clone of the hepatitis C virus genotype 1 prototype sequence. J Gen Virol. 2001;82:1291-1297. [PubMed] |
| 19. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2294] [Cited by in RCA: 2251] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 20. | Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001-13014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 980] [Cited by in RCA: 1024] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 21. | Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3' end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 292] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 22. | Bartenschlager R, Frese M, Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res. 2004;63:71-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 213] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 23. | Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci USA. 2004;101:13038-13043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 255] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 24. | Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, Bartholomew L, De Francesco R. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J Virol. 2004;78:13306-13314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002;76:2997-3006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 329] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 26. | Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci USA. 2002;99:14416-14421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 200] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 27. | Yi M, Bodola F, Lemon SM. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology. 2002;304:197-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 82] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2323] [Cited by in RCA: 2275] [Article Influence: 113.8] [Reference Citation Analysis (0)] |
| 29. | Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64:334-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 188] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 30. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1850] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 31. | Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci USA. 2006;103:7408-7413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 602] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 32. | Kalinina O, Norder H, Mukomolov S, Magnius LO. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol. 2002;76:4034-4043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 240] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 33. | Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310-2315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 309] [Cited by in RCA: 301] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 34. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1475] [Cited by in RCA: 1467] [Article Influence: 73.4] [Reference Citation Analysis (0)] |
| 35. | Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689-2699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 736] [Cited by in RCA: 724] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 36. | Windisch MP, Frese M, Kaul A, Trippler M, Lohmann V, Bartenschlager R. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J Virol. 2005;79:13778-13793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |