Published online Apr 21, 2007. doi: 10.3748/wjg.v13.i15.2218
Revised: January 15, 2007
Accepted: February 8, 2007
Published online: April 21, 2007
AIM: To study the gene expression changes in pancreatic cystic neoplasm in SV40Tag transgenic mice model and to provide information about the prevention, clinical diagnosis and therapy of pancreatic cancer.
METHODS: Using the pBC-SV40Tag transgenic mice model of pancreatic cystic neoplasm, we studied the gene expression changes by applying high-density microarrays. Validation of part gene expression profiling data was performed using real-time PCR.
RESULTS: By using high-density oligonucleotide microarray, of 14 113 genes, 453 were increased and 760 decreased in pancreatic cystic neoplasm, including oncogenes, cell-cycle-related genes, signal transduction-related genes, skeleton-related genes and metabolism-related genes. Among these, we confirmed the changes in Igf, Shh and Wnt signal pathways with real-time PCR. The results of real-time PCR showed similar expression changes in gene chip.
CONCLUSION: all the altered expression genes are associated with cell cycle, DNA damage and repair, signal pathway, and metabolism. SV40Tag may cooperate with several proteins in promoting tumorigenesis.
- Citation: Feng J, Sun Q, Gao C, Dong J, Wei XL, Xing H, Li HD. Gene expression analysis of pancreatic cystic neoplasm in SV40Tag transgenic mice model. World J Gastroenterol 2007; 13(15): 2218-2222
- URL: https://www.wjgnet.com/1007-9327/full/v13/i15/2218.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i15.2218
Pancreatic cystic neoplasms comprise a heterogeneous group of pathologic entities. Although the incidence rate and lethality of the pancreatic cystic neoplasm is not as high as that of the pancreatic adenocarcinoma, some of the cystic pancreatic neoplasms can be developed into metastatic pancreatic adenocarcinoma. Due to the low incidence and recent nosographic arrangement, the natural evolution and the clinical development of pancreatic cystic neoplasms are not well known. It is, therefore, difficult to identify all types of these cystic lesions. Pancreatic cancers were rarely observed spontaneously or induced by carcinogen administration in the laboratory mice. However, genetic engineering has been shown to effectively generate mice with germline oncogenic lesions similar to human pancreatic adenocarcinomas[1].
We have successfully generated a transgenic animal model with classic features of pancreatic cystic neoplasm[2]. This transgenic mice model formed pancreatic cystic neoplasms owing to the expression of SV40Tag. In order to identify the tumor-associated genes of pancreatic cystic neoplasm, we analyzed the gene expression changes in this tumor by using gene chip technique. To further confirm the gene expression profiling data from gene chip experiment, some of the genes with high-mutation in cancer were validated by real-time PCR. This study may help to search the genes associated to pancreatic cystic neoplasm; and it will provide some information about the prevention, clinical diagnosis and treatment of pancreatic cystic neoplasm.
pBC-SV40Tag transgenic mice model of pancreatic cystic neoplasm and FVB mice were provided by Shanghai Institute of Brain Functional Genomics, East China Normal University. Mice were maintained in standard laboratory conditions on a 12 h: 12 h light-dark cycle. We randomly selected three pBC-SV40Tag transgenic mice and three normal mice, dissected tumor and normal pancreas, and separated them into two independent tissue pools for the extraction of poly (A) mRNA. Total RNA was extracted and reverse transcribed into cDNA using the SuperScript First-Strand Synthesis System for RT-PCR (Life Technologies, INVITROGEN, Carlsbad, CA). The high-density oligonucleotide microarray (gene chip) analysis was conducted as previously described[3,4]. Microarray experiments were performed using an Agilent G4121A Mouse Oligo Microarray Kit following the manufacturer’s instructions. Data were analyzed by Imagene software and normalized by Feature Extraction. To ensure the reliability of the data, we conducted hybridization experiments in duplicates consisting of two independent mRNAs and two sets of duplicate microarrays. In agreement with our previous experience with DNA microarrays, we found our procedures involving duplicate experiments provided consistent and reproducible hybridization signals.
The expression profiling of several genes involved in signal pathways identified using gene chip were confirmed by fluorescence quantitative real-time PCR. The experiment was performed on the Option 2 (MJ research, USA). PCR primers were designed using primer 5.0. PCR primers were as follows: igf-ir: 5-GCCTTGGTCTCCTTGTCCTT-3′ (forward) and 5′-CTCTGGCGTCCCTTGGTT-3′ (reverse); igf-ii: 5′-AGCCGTGGCATCGTGGAA-3′ (forward) and 5′-GCGGGGTCTTTGGGTGGT-3′ (reverse); irs-1: 5′-CCTGGAGTATTATGAGAACGAG-3′ (forward) and 5′-CGGCAATGGCAAAGTGT-3′ (reverse); igfbp-1: 5′-GCCCGAGTTCCTAACTGTTG-3′ (forward) and 5′-CAGCAGCCTTTGCCTCTTC-3′ (reverse); igfbp-2: 5′-CCAGACGCTACGCTGCTATC-3′ (forward) and 5′-CACTGCTACCACCTCCCAAC-3′ (reverse); igfbp-3: 5′-GCAGCCTAAGCACCTACCT-3′ (forward) and 5′-CCTCTGGGACTCAGCACAT-3′ (reverse); igfbp-4: 5′-GGTTGCGAGGAGTTGGTG-3′ (forward) and 5′-GTGGGTACGGCTCTGTGAG-3′ (reverse); igfbp-5: 5′-TGAGATGAGACAGGAATCCGAACA-3′ (forward) and 5′-CGAAGGCGTGGCACTGAAAG-3′ (reverse); igfbp-6: 5′-CCGTCGGAGGAGACTACAAAG-3′ (forward) and 5′-CCATCTGGAGACACTGGCAAA-3′ (reverse); β-catenin: 5′-TCTACGCCATCACGACAC-3′ (forward) and 5′-CAGACAGACAGCACCTTC-3′ (reverse); shh: 5′-CGGCAGATATGAAGGGAAGA-3′ (forward) and 5′-GGTCCAGGAAGGTGAGGAAGT-3′ (reverse); gli2: 5′-CACAGGGCGGGCACAAGAT-3′ (forward) and 5′-GGAGGGCAGTGTCAAGGAA-3′ (reverse); sufu: 5′-TCCAGGTTACCGCTATCGTC-3′ (forward) and 5′-TCCACTGTTGGGCTGAATGT-3′ (reverse); and GAPDH: 5′-TCAACGACCCCTTCATTGAC-3′ (forward) and 5′-ATGCAGGGATGATGTTCTGG-3′ (reverse). GAPDH was used as a reference gene. PCR reaction was performed using 20 μL of total reaction mixture volume, containing 1 μL of cDNA reaction products and 0.2 μL of SYBR Green I as follows: one cycle at 95°C for 10 min, followed by 40 amplification cycles, each cycle consisting of denaturation at 95°C for 30 s, primer annealing at 59°C for 30 s and extension at 72°C for 30 s. Fold change was determined based on average cycle threshold (Ct) values for duplicates. Gene expression levels were calculated and presented with 2-ΔΔCt values[5].
The RNA electrophoresis revealed two clear and bright bands, which correspond to 28 S and 18 S (Figure 1). Spectrophotometer measure showed absorption of total RNA at A260/A280 greater than 1.9.
Gene chip analysis focused on genes altered by 3.0- or 0.3-fold change with a P-value ≤ 0.05. Compared to the tumor and normal pancreas, of 1 4113 genes, 552 were increased in pancreatic cystic neoplasm, such as replication factor C (recc1) and n-myc downstream-regulated 3 (ndr3), and 759 were decreased in pancreatic cystic neoplasm, such as bcl2-like 2 (bcl2l2) and programmed cell death 2 (pdcd2). All the altered expression genes were associated with cell cycle, DNA damage and repair, signal pathway, metabolism and so on (Table 1). According to the gene function, about 26% were associated with signal transduction, and 8.4% with cell cycle.
| Category | Gene name | GenBank ID | Average ratio |
| Signal transduction | RB-associated KRAB repressor (rbak) | AF226870 | 2.373022 |
| Regulator of G-protein signaling 2 (rgs2) | NM_009061 | 9.531784 | |
| G-protein-coupled receptor 65 (gpcr65) | NM_008152 | 3.032290 | |
| G protein-coupled receptor 19 (gpr19) | NM_008157 | 3.761271 | |
| Sonic hedgehog (shh) | X76290 | 0.366694 | |
| Nerve growth factor receptor (ngfr) | NM_033217 | 0.322626 | |
| Platelet-derived growth factor receptor, alpha polypeptide (pdgfra) | NM_011058 | 2.925733 | |
| Cholecystokinin A receptor (cckar) | NM_009827 | 2.359371 | |
| Oncogene | Kit oncogene (kit) | NM_021099 | 5.686938 |
| Myeloblastosis oncogene (myb) | NM_033597 | 2.834637 | |
| C-mer proto-oncogene (mer) | NM_008587 | 3.447795 | |
| Cell cycle | Cyclin A2 (ccna2) | NM_009828 | 9.783554 |
| Cyclin G2 (ccng2) | NM_007635 | 2.630338 | |
| Cyclin-dependent kinase inhibitor 1C (cdkn1c) | NM_009876 | 2.802018 | |
| Cell division cycle 25 homolog C (cdc25c) | NM_009860 | 3.383045 | |
| Cyclin-dependent kinase inhibitor 2D (cdkn2d) | NM_009878 | 4.352507 | |
| Adducin 1 (alpha) (add1) | NM_013457 | 2.672981 | |
| Cytoskeleton | Tubulin beta 5 (tubb5) | NM_011655 | 2.867130 |
| Matrix metalloproteinase 19 (mmp19) | NM_021412 | 0.509155 | |
| Capping protein (actin filament), gelsolin-like (capg) | NM_007599 | 2.840273 | |
| Metabolism | InsulinI(ins1) | NM_008386 | 0.573778 |
| InsulinII(ins2) | NM_008387 | 0.211135 | |
| Acylglycerol-3-phosphate O-acyltransferase 3 (agpat3) | NM_053014 | 2.607845 | |
| Carbonic anhydrase 2 (car2) | NM_009801 | 2.452762 | |
| Fatty acid-coenzyme A ligase, long chain 4 (facl4) | NM_019477 | 3.001425 | |
| Phosphofructokinase, platelet (pfkp) | NM_019703 | 2.690855 | |
| Apoptosis | Death-associated protein (dap) | AK014991 | 0.358995 |
| Caspase recruitment domain family, member 15 (card15) | AK089843 | 0.329593 | |
| Cell death-inducing DNA fragmentation factor, alpha subunit-like effector B (cideb) | NM_009894 | 0.221076 | |
| Mus musculus Bcl2-like 2 (bcl2l2) | NM_007537 | 0.784750 | |
| Tumor necrosis factor receptor superfamily, member 10b (tnfrsf10b) | NM_020275 | 0.451223 | |
| Programmed cell death 2 (pdcd2) | NM_008799 | 0.481340 | |
| Cell antigen | CD68 antigen (cd68) | NM_009853 | 3.388384 |
| Centromere autoantigen C (cenpc) | NM_007683 | 2.372337 |
Several oncogenes were obviously up-regulated in the pancreatic cystic neoplasm, such as kit oncogene, myeloblastosis oncogene (myb). Members of Wnt receptor pathway, β-catenin interacting protein 1 (catnbip1) and Wnt1 inducible signaling pathway protein 2 (wisp2) were also up-regulated. Gpcr65, 19, members of G-protein-coupled receptors family, were found to be changed in the tumor. Several genes involved in apoptosis, such as bcl2-like 2 (bcl2l2), programmed cell death 2 (pdcd2), were down-regulated (Table 1).
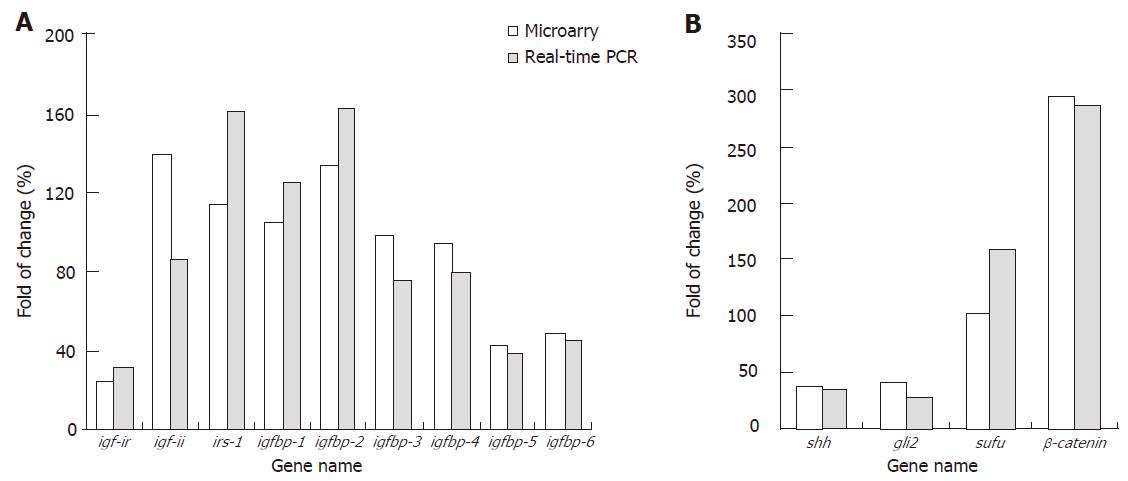
Interestingly, several members of Igf pathway activated in many tumors, such as insulin-like growth factor (igf-i), igf-i receptor (igf-ir) and insulin-like receptor substrateI(irs-1), were down-regulated or showed a little change in the tumor (Figure 2); but insulinI(ins1) and insulin II (ins2), two hormones of Igf signal pathway and involved in glucose transport, were down-regulated in pancreatic cystic neoplasm. Sonic hedgehog (shh) and gli2, involved in the development and cell-cell signaling, were also down-regulated in pancreatic cystic neoplasm.
We performed real-time PCR to further confirm the gene expression profiling data using gene chip. The results demonstrated that most of the chosen genes showed similar expression changes in both gene chip and real-time PCR (Figure 2). The data also revealed that the fold changes of most chosen genes were higher in real-time PCR than in gene chip. Thus, we realized that the real-time PCR method is more sensitive and reliable than microarray technology.
With the completion of Human Genome Project and beginning of proteomics, it is very necessary to study the gene function. Cancer is caused by the accumulation of genetic mutations, such as oncogenes or tumor suppressor genes. Gene chip has become one of the most important technologies since 1990’s. It has been widely used to study the gene expression analysis, tumor classification and cancer progression. It has shown enormous potential in gene function research, clinical diagnosis, and drug screening[6].
In this study, we analyzed the gene expression changes in pBC-SV40Tag transgenic mice model by applying high-density microarrays. Validation of the gene expression profiling data was performed by using real-time PCR. We found obvious changes in genes were involved in signaling pathway, metabolism and cell cycle.
Igf-ir is a multifunctional tyrosine kinase involved in normal and pathological growth of cell[7-9]. It can support malignant growth of cell by stimulating cell proliferation, protecting cells from apoptosis, triggering a transformed phenotype when the receptor molecular is grossly over-expressed[9-12]. Irs-1 is a major cytosolic signaling molecule for igf-ir[13]. But it is worth noting that they were down-regulated or had a little change in our study, and the expression of other members, such as insulinI(ins1) and ins2, were also decreased. It is consistent with literature that the majority of mice with pancreatic cystic neoplasm have diabetes or high blood sugar. It suggests that pancreatic lesion in pBC-SV40Tag transgenic mice may affect the secreting function of pancreatic cells. Igf signalling pathway does not promote the pancreatic cystic neoplasm.
The Hedgehog signaling pathway is known to play a very important role during development, regulation of both cell fate and proliferation[14,15]. The Shh signaling pathway is required in many mammalian tissues for embryonic patterning, cell proliferation and differentiation. In addition, inappropriate activation of the pathway has been implicated in many tumors[16,17]. Gli2, the members of Shh pathway, plays key roles in activating the signal pathway, and promoting gene expression. But in our study, shh and gli2 showed low expression level in pBC-SV40Tag transgenic mice, which might be the cooperative effects of several other signal pathways.
Wnt pathway can be detected in many tumors. The critical mediator, β-catenin, is an important downstream protein of it. It initiates a complex signaling cascade that plays an important role in regulating cell proliferation and differentiation. It is a member of cytoskeleton, and also involved in signal transduction[18]. It also plays important roles in DNA binding and transcription. Moreover, it can cooperate with the nuclear protein, such as cyclin D1, c-myc, to promote the gene expression. In our study, β-catenin, cyclin D1, β-catenin-interacting protein 1 (catnbip1) and WNT1-inducible signaling pathway protein 2 (wisp2) were also up-regulated obviously. It may be an important clue for the nosogenesis study of pBC-SV40Tag transgenic mice.
Recently, the studies on G protein-coupled receptors (GPCRs) have shown that GPCRs modulate diverse physiological and behavioral signaling pathways by virtue of changes in receptor activation and inactivation states. We found several changes of GPCRs and the regulators in pBC-SV40Tag transgenic mice by using microarray, such as gpcr65, gpr19. They both were up-regulated in pancreatic cystic neoplasm, but we could not find the correlative reports. It suggests us they may promote the tumorigenesis in the transgenic mice model.
On the basis that SV40Tag is a viral oncogene, epidemiology studies have shown that SV40Tag can be detected in several human tumors, such as lymphoma, brain tumor and bone tumor[19-21]. Furthermore, SV40 genomic DNA sequence can be detected from tumors[22]. So, we speculated that Wnt pathway and GPCRs family may cooperate with SV40Tag to promote pancreatic tumorigenesis. About 98% of the 94-ku phosphorylated SV40Tag locates in the nucleus. Several in vitro studies have demonstrated that SV40Tag can integrate into cell genome, destroy its stability and activate the abnormal gene expression[23-25]. In pBC-SV40Tag transgenic mice model, SV40Tag may have the biological activity similar to that in vitro. We speculate that it has the ability to translocate some proteins from plasma to nucleus, further activate the downstream gene expression, and thereby promoting the tumorigenesis.
To study the nosogenesis of SV40Tag and the related proteins in the transgenic mice, we should further apply immunohistochemistry, immunoprecipitation experiment to confirm our hypotheses. Hence, we should further study some of the strategies that are being used or can be explored to target the components of these signaling pathways in drug discovery of pancreatic cystic neoplasm.
In conclusion, we studied the gene expression changes by applying high-density microarrays. Validation of the gene expression profiling data was performed using real-time PCR. Gene chip may be a powerful strategy to identify the cancer-associated genes. The data suggests that SV40Tag may cooperate with members of Wnt pathways, GPCRs and play important role in the pancreatic cystic neoplasm. We speculate that they may cooperate with other signal pathways in promoting tumorigenesis. All these may provide some information about the prevention, clinical diagnosis and treatment of pancreatic cystic neoplasm.
S- Editor Wang J L- Editor Kumar M E- Editor Che YB
| 1. | Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112-3126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 772] [Cited by in RCA: 816] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 2. | Sun Q, Feng J, Wei XL, Zhang R, Dong SZ, Shen Q, Dong J, Li HD, Hu YH. Generation and characterization of a transgenic mouse model for pancreatic cancer. World J Gastroenterol. 2006;12:2785-2788. [PubMed] |
| 3. | Mody M, Cao Y, Cui Z, Tay KY, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien JZ. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci USA. 2001;98:8862-8867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 157] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 4. | Rampon C, Jiang CH, Dong H, Tang YP, Lockhart DJ, Schultz PG, Tsien JZ, Hu Y. Effects of environmental enrichment on gene expression in the brain. Proc Natl Acad Sci USA. 2000;97:12880-12884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 452] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 5. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133878] [Article Influence: 5578.3] [Reference Citation Analysis (1)] |
| 6. | Kim DS, Watkinson JC. Gene chip expression analysis in head and neck cancer. Clin Otolaryngol Allied Sci. 2002;27:296-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 7. | Baserga R, Resnicoff M, Dews M. The IGF-I receptor and cancer. Endocrine. 1997;7:99-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Baserga R. The IGF-I receptor in cancer research. Exp Cell Res. 1999;253:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 212] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 9. | Reiss K, Valentinis B, Tu X, Xu SQ, Baserga R. Molecular markers of IGF-I-mediated mitogenesis. Exp Cell Res. 1998;242:361-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Baserga R, Sell C, Porcu P, Rubini M. The role of the IGF-I receptor in the growth and transformation of mammalian cells. Cell Prolif. 1994;27:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 99] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Valentinis B, Morrione A, Peruzzi F, Prisco M, Reiss K, Baserga R. Anti-apoptotic signaling of the IGF-I receptor in fibroblasts following loss of matrix adhesion. Oncogene. 1999;18:1827-1836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 68] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Morrione A, DeAngelis T, Baserga R. Failure of the bovine papillomavirus to transform mouse embryo fibroblasts with a targeted disruption of the insulin-like growth factor I receptor genes. J Virol. 1995;69:5300-5303. [PubMed] |
| 13. | White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998;182:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 512] [Cited by in RCA: 472] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 14. | Hammerschmidt M, Brook A, McMahon AP. The world according to hedgehog. Trends Genet. 1997;13:14-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 441] [Cited by in RCA: 418] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 15. | Goodrich LV, Scott MP. Hedgehog and patched in neural development and disease. Neuron. 1998;21:1243-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 195] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 16. | Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, Seuanez HN, O'Brien SJ, Vogelstein B. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104-3113. [PubMed] |
| 17. | Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 372] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 18. | Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1521] [Cited by in RCA: 1529] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 19. | Cristaudo A, Vivaldi A, Sensales G, Guglielmi G, Ciancia E, Elisei R, Ottenga F. Molecular biology studies on mesothelioma tumor samples: preliminary data on H-ras, p21, and SV40. J Environ Pathol Toxicol Oncol. 1995;14:29-34. [PubMed] |
| 20. | Pass HI, Kennedy RC, Carbone M. Evidence for and implications of SV40-like sequences in human mesotheliomas. Important Adv Oncol. 1996;89-108. [PubMed] |
| 21. | Carbone M, Rizzo P, Procopio A, Giuliano M, Pass HI, Gebhardt MC, Mangham C, Hansen M, Malkin DF, Bushart G. SV40-like sequences in human bone tumors. Oncogene. 1996;13:527-535. [PubMed] |
| 22. | Bergsagel DJ, Finegold MJ, Butel JS, Kupsky WJ, Garcea RL. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N Engl J Med. 1992;326:988-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 274] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 23. | DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1039] [Cited by in RCA: 1090] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 24. | Ludlow JW, DeCaprio JA, Huang CM, Lee WH, Paucha E, Livingston DM. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell. 1989;56:57-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 366] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 25. | Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 407] [Article Influence: 12.3] [Reference Citation Analysis (0)] |