Published online Mar 21, 2007. doi: 10.3748/wjg.v13.i11.1687
Revised: February 23, 2006
Accepted: March 14, 2007
Published online: March 21, 2007
AIM: To study susceptibility genes which may play a potential role in the pathogenesis and etiology of inflammatory bowel disease (IBD).
METHODS: To identify potential susceptibility genes we performed global gene expression profiling in patients with IBD and control specimens. For determination of an intrinsic gene expression profile in ulcerative colitis (UC) and Crohn's disease (CD) compared to normal subjects, mucosal biopsies of non-inflamed regions of the colon and the terminal ileum were subjected to DNA microarray analysis. Real-time RT-PCR and immunohistochemistry were used for verification of selected regulated candidate genes and a genetic analysis was performed.
RESULTS: We could show that aquaporin-8 (AQP8) mRNA and protein levels were significantly increased in the colon of UC patients compared to controls. Genetic analysis of the six exons and the promoter region of AQP8, however, revealed no mutations or polymorphisms in IBD patients.
CONCLUSION: Our results suggest that upregulation of AQP8 in the colon of UC patients represents a secondary phenomenon which may, due to altered water exchange of the distal intestinal mucosa, disturb the physiologic colonic mucus barrier and thus lead to chronic infla-mmation and ulceration.
- Citation: Zahn A, Moehle C, Langmann T, Ehehalt R, Autschbach F, Stremmel W, Schmitz G. Aquaporin-8 expression is reduced in ileum and induced in colon of patients with ulcerative colitis. World J Gastroenterol 2007; 13(11): 1687-1695
- URL: https://www.wjgnet.com/1007-9327/full/v13/i11/1687.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i11.1687
Crohn’s disease (CD) and Ulcerative colitis (UC) are inflammatory bowel diseases (IBD) with shared clinical and demographic features. Although, there has been progress in understanding the pathogenesis of these diseases in the last decade, the etiology remains unknown. The current hypothesis suggests that environmental factors trigger a breakdown in the regulatory constraints on mucosal immune responses to enteric bacteria in genetically susceptible individuals[1]. Mapping studies indicate a strong inherited component but a large number of putative susceptibility loci has complicated the identification of IBD genes. So far, several potential IBD susceptibility loci have been identified in different genomic regions, containing numerous putative candidate genes[2,3]. In the absence of a priori candidate genes, experimental techniques like quantitative expression studies, such as microarray technology, seem to be an useful approach to genome-wide searches for IBD genes and to narrow down the number of genes to a reasonable size[4,5]. Reproducible results were obtained by Dieckgraefe et al[5] who studied differential gene expression in UC mucosal specimens exclusively, using the first generation of Affymetrix arrays (Hum 6000, a set of four chips which contain 256 000 individual oligonucleotide features representing 6500 human genes and Expressed Sequence Tags (ESTs), and by Lawrance et al[4] who used the second generation of Affymetrix arrays (HuGene Fl arrays 900160 and 900183) to screen samples from both CD and UC patients. By combining information from published IBD linkage analysis and association studies with our Affymetrix microarray results (mapping and arraying strategy[6]), we obtained several genes of interest. Among them, we found the significantly differentially regulated aquaporin-8 (AQP8) gene, which is located on chromosome 16p12.1 and thus within the IBD locus 8[7]. Therefore, we focused on the analysis of this gene in the present study.
The aquaporins (AQPs) are a family of small (about 30 kDa) integral membrane proteins that function as water channels in animals, plants and bacteria. So far, 13 AQP homologues have been identified in mammals and an increasing number of disturbances have been found associated to the abnormal function of these proteins[8-11]. Among mammalian aquaporins, two subgroups have been defined: “aquaporins” and “aquaglyceroporins”. Compared with the sequence of the aquaporins (e.g. AQP1, AQP2, AQP4, AQP5, AQP6, and AQP8), the aquaglyceroporins (e.g. AQP3, AQP7, and AQP9) contain two additional peptide spans required for the transport of glycerol, urea and even larger solutes[9,11]. The capability to transport glycerol as backbone molecule for triglycerides also links aquaporins to lipid metabolism, thus regional expression of AQPs may influence fatty acid metabolism in highly asorptive tissues such as the intestine.
Due to its cellular and subcellular distribution it is concluded that AQP8 plays an important role in the absorption of water in the intestine and that AQP8 may also be involved in processes of intracellular osmoregulation and mucosal fluid fluxes[12].
Moreover, Fischer et al[13] assume that AQP8 is a marker of normal proliferating colonic epithelial cells because they failed to detect AQP8 mRNA in de-differentiated colonic cells during colorectal carcinogenesis. Taken together, literature data underscore the importance of AQP8 in the absorption of fluids in the gastrointestinal (GI) tract and putatively, in intracellular osmoregulation.
In this study we have employed microarray experiments to screen mucosal gene expression in non-inflamed regions (10 cm distant from pathological areas, no endoscopic signs of inflammation) of UC and CD patients and healthy controls from two different locations, terminal ileum and transverse colon. Therefore, we used the Affymetrix Human Genome U133A and U133B Gene Chip set which covers 39 000 transcripts and variants, including more than 33 000 well-substantiated human genes. The microarray experiments were performed and analyzed according to the Gene Chip Expression Analysis Technical Manual (Affymetrix, Santa Clara, USA). The complete data set is publicly available in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) through the accession number GSE1152. To correct for interindividual differences and to enrich for IBD-specific transcriptional events, we pooled four samples from each RNA source: UC patients, CD patients and controls, terminal ileum and transverse colon, respectively. Patients’ characteristics are given in Table 1. Only genes whose expression differed by a factor ≥ 2 were considered as significantly regulated and were included in the further analyses. For pooling, 2.5 μg of total RNA from each sample were used. This approach allowed to preserve remaining RNA material, which can be used for real-time RT-PCR evaluation of significant and interesting gene candidates[14]. The study was approved by the institutional ethics committee of the University of Regensburg.
| Disease | Controls | CD | UC |
| n | 4 | 4 | 4 |
| Sex (M/F) | 1/3 | 1/3 | 2/2 |
| Age, range (yr) | 52-60 | 23-43 | 20-48 |
| Age, mean (yr) | 56 | 32 | 35 |
In the first step, a complete search for known IBD loci in the Online Mendelian Inheritance in Man (OMIM) database, containing a detailed description of all already IBD linked susceptibility loci including the connected sequence-tagged-site (STS) markers has been performed. These markers were compared to the Human Genome Project Working Draft (UCSC) database to identify all known genes located in the particular chromosomal regions. Based on a critical survey of known databases, reference sequences were retrieved and a complete list including mRNA sequences with Unigene numbers was assembled for these putative IBD candidate genes. Finally, based on Unigene IDs, it was possible to determine the expression patterns of all transcripts present on the Affymetrix U133A and U133B Gene Chips located in IBD candidate genes.
RNA extraction: Biopsy samples of 4 patients with UC, 4 patients with CD, and of 4 control patients (Table 1), who underwent colonoscopy for other reasons than IBD, including tumor staging, were obtained from ileum and transverse colon, respectively and manually homogenized in liquid nitrogen by a pestle. Afterwards, RNA extraction was carried out according to the manufacturer's instructions using the RNeasy Midi Kit (Qiagen, Hilden, Germany). The purity and integrity of the RNA were assessed on the Agilent 2100 bioanalyzer with the RNA 6000 Nano LabChip reagent set (Agilent Technologies, USA). The RNA was quantified spectrophotometrically and then stored at -80°C.
cDNA synthesis: First-strand cDNA synthesis was performed with the Reverse Transcription System from Promega. To a master mixture (prepared in house) containing 5 mmol/Lol/L MgCl2, 1x reverse transcription buffer, 1 mmol/L deoxynucleotide triphosphate mixture, 1 unit/µL recombinant RNasin® ribonuclease inhibitor, 0.75 U/μL AMV reverse transcriptase and 1 μg of random hexamer primers, we added 2 μg of total RNA and sterile H2O to a final volume of 40 μL. The reaction mixture was incubated at 42°C for 60 min, followed by heat inactivation of the enzyme at 95°C for 5 min. After cooling on ice for 5 min, the cDNA was stored at -20°C.
TaqMan primers and probe design: The mRNA sequence of the human AQP8 was derived from the NCBI Nucleotide database (accession number NM_001169) and primers and TaqMan probes were designed with PrimerExpress Software, version 2.0 (Applied Biosystems). All oligonucleotides and 6-carboxyfluorescein (FAM)-labeled probes for TaqMan expression analysis were obtained from MWG-Biotech (AQP8 forward primer: 5’-gcctgaatttggcaatgaca-3’; AQP8 probe: 5’- cagggagccgagcgtgggtg-3’; AQP8 reverse primer: 5’-aaaccgttcgtaccaggacact-3’). For the normalization of our results, we used a VICTM-labeled glyceraldehyde 3-phosphate dehydrogenase (GAPDH) TaqMan PDAR endogenous control reagent set (Applied Biosystems). Each of the probes was quenched by 6-carboxytetramethylrhodamine (TAMRA) at its 3’ end.
Generation of calibration curves and TaqMan real-time RT-PCR: To quantify the results obtained by real-time RT-PCR, we used a calibration curve. For this purpose, from a stock solution of HT29 cDNA generated from total RNA, serial dilutions with 50, 25, 12.5, and 6.25 ng of cDNA were prepared. TaqMan PCR assays were performed on an ABI Prism 7900 HT Sequence Detection System (Perkin-Elmer Applied Biosystems). For quantification of the AQP8 gene, we prepared a master mixture containing 10 μL of 2 × TaqMan Universal PCR Master Mix, 1 μL of both gene-specific forward and reverse primer (each at 18 μmol/L), respectively, 1 μL of the gene-specific probe (5 μmol/L) and 2 μL of sterile water and aliquoted it into the wells of a 384-well optical plate. A master mixture for the endogenous control GAPDH containing 10 μL of 2 × TaqMan Universal PCR Master Mix, 1 μL of predeveloped TaqMan assay reagents (PDAR) from the endogenous control reagent set and 4 μL sterile water was treated similarly. Finally, triplicates of cDNA templates equivalent to 50 ng of RNA were added to a final volume of 20 μL. The thermal cycling conditions were 2 min at 50°C and 10 min at 95°C followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. For data analysis the Sequence Detector Software SDS 2.0 (Applied Biosystems) was used and the analysis was performed as previously described by Langmann et al[15].
Immunohistochemical stainings for AQP8 were performed on 2 μm sections of routinely processed, formalin-fixed (4% neutral-buffered formaldehyde), paraffin-embedded tissues using the alkaline phosphatase anti-alkaline phosphatase technique (APAAP). After dewaxing, the slides were incubated for 10 min at 90°C in Target Unmasking Fluid (TUFTM, Monosan, distributed by Biozol, Eching, Germany) and washed with distilled water and Tris-buffered saline (TBS; pH 7.6). The sections were incubated for 48 h at 4°C with the primary antibody (rabbit polyclonal anti-AQP8; Chemicon, Temecula, CA, USA), diluted 1:20 in TBS/0.2% acetylated bovine serum albumin (BSA; Aurion, Wageningen, Netherlands)/0.1% Tween-20 (Carl Roth GmbH, Karlsruhe, Germany) with addition of 2.5 mg/mL normal human immunoglobulins (γ-venin; Behring, Marburg, Germany). After washing with TBS/0.2% BSA/0.1% Tween-20, polyclonal mouse anti-rabit antibodies were added for 30 min (dilution 1:50; Dako, Hamburg, Germany), followed by rabbit-anti mouse bridging antibodies (1:25; Dako) and the APAAP-complex (1:50; Dako), which were applied in two separate incubation cycles of 30 and 15 min, respectively. Naphthol AS-biphosphate (Sigma) and new fuchsin (Merck, Darmstadt, Germany) served as the substrate for alkaline phosphatase. Finally, the sections were counterstained with hematoxylin and mounted (Aquatex, Merck, Darmstadt, Germany). Incubations with use of normal rabbit immunoglobulins (Dako) instead of the AQP8 specific antibody as the primary reagent served as negative controls.
DNA isolation: Genomic DNA was extracted from whole blood EDTA samples using QIAgen Midi Kit. For single samples, the quality of the DNA was checked by agarose gel and the concentration was determined by absorption measurement. For larger sets of samples, the DNA was pipetted into microtiter plates, the concentration was determined using a Picogreen Assay (Molecular Probes) and the DNA was normalized to a concentration of 10 ng/μL. Normalization was carried out using BFX-Normalization Software and Biomek FX pipetting robot (Beckman Coulter).
DNA sequencing: To identify genetic variants in the AQP8 gene DNA sequencing of the promoter region and its six exons was performed on an ABI Prism 3100 Genetic Analyser with Big Dye Terminator technology in 100 patients with either CD or UC and in 50 healthy controls. PCR products were generated with the set of primers presented in Table 2. The Sequencher software (GeneCodes) was used to align the trimmed sequences to each other or to the wild type sequence. All electronically reported abnormalities and heterozygosities were checked individually in the electropherogram.
| AQP8 fragment | Primer designation | 5’-3’ sequence |
| Promotor region | Pro-forward | CCGTGTTAGCCAGGATGG |
| Pro-reverse | GAAACCTGCACCTGCTGTG | |
| Exon 1 | Ex1-forward | CCCTGCCCTGTTGAGATTTA |
| Ex1-reverse | AGGGAAAAAGGGACAGAGGA | |
| Exon 2 | Ex2-forward | AAGAGTCCGATGTTTGTGCC |
| Ex2-reverse | CTTTGCTTTCCACACCCAGT | |
| Exon 3 | Ex3-forward | ACACTGTCTCAAGTGCCAGC |
| Ex3-reverse | CACACCCACATATGCACCTC | |
| Exon 4 | Ex4-forward | GCAGGGTCGCACAGTAAAAT |
| Ex4-reverse | CTGGCCCCTAATAGCAACTG | |
| Exon 5 | Ex5-forward | AGCCCCTCTGCCTTCTTTAG |
| Ex5-reverse | TTCCAAACCCAAGTGAGAGC | |
| Exon 6 | Ex6-forward | AGCCTGGAGACATGACGAAG |
| Ex6-reverse | AGAGCCTCCTCAGCAGTCAG |
TaqMan allelic discrimination: As a genetic case-control association study TaqMan allelic discrimination was performed in 220 patients with UC, in 181 patients with CD, and in 250 healthy controls to obtain single nucleotide polymorphism (SNP) frequencies in IBD patients compared to controls. In brief, this TaqMan assay involves the use of two detection probes, each recognizing a specific allele. Each probe is labeled with a fluorophore on its 5’ end (vic or 6-fam) and a non-fluorescent quencher (nfq) attached to a minor-groove-binder (mgb) on its 3’ end. The minor-groove-binder increases the melting temperature of the probe thus allowing a more stringent annealing. The nfq quenches the fluorescence of the flurophore as long as both are attached to the same oligonucleotide. During PCR, a probe with a perfect match will be cut into pieces by the 5’-3’ exonuclease activity of the Taq polymerase releasing unquenched fluorphore, while a mismatched probe will be displaced. End-point fluorescence intensity was measured in an ABI7900HT machine (Applied Biosystems) and analyzed by using the ABI Sequence Detector (SDS) Software version 2.0 (Applied Biosystems). Sequence specific TaqMan primers and probes were designed by using the PrimerExpress Software, version 2.0 (Applied Biosystems) and are presented in Table 3. Standard ABI protocol for TaqMan SNP analysis was used.
| AQP8 SNP | Primer designation | 5’-3’ Sequence |
| AQP8-A212T | Forward primer | CTTGGTGCCTGGGTGTTTG |
| Reverse primer | AGCCAGTAGATCCAGTGGAAGTTC | |
| FAM-Probe | CATGAATCCCACCCGT | |
| VIC-Probe | TCCCGCCCGTGCT | |
| AQP8-A260P | Forward primer | GAGATGGGAAGACCCGCC |
| Reverse primer | GGAATCCCACGAGCTCTGC | |
| FAM-Probe | TCCTGAAGGCTCGGTGA | |
| VIC-Probe | TCCTGAAGCCTCGGTGA | |
| AQP8-R261Q | Forward primer | GGCAGGAAATGCAGGAACTC |
| Reverse primer | GGGAAGACCCGCCTCATC | |
| FAM-Probe | AGCTCTGCTTCACTGAG | |
| VIC-Probe | AGTTCTGCTTCACCGAG |
All data are expressed as the mean ± standard deviation. Statistical significance was determined by student’s t-test for unpaired samples. Frequency table and χ² test were performed to analyze allelic discrimination data. For both statistical procedures a value of P < 0.05 was considered statistically significant.
In this study the expression of more than 33 000 well-substantiated human genes in the complex diseases CD and UC was analyzed by a microarray based system using Affymetrix Human Genome U133A and U133B Gene Chips. To identify putative IBD candidate genes within the large number of known genes, we combined information from published IBD linkage analysis and association studies with our Affymetrix microarray results.
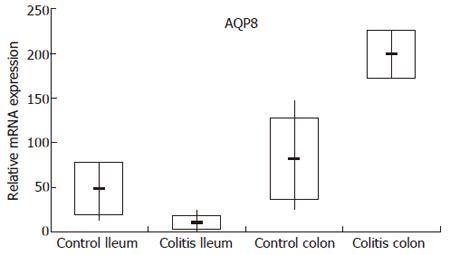
Table 4 gives an interesting selection of differentially regulated genes located in IBD candidate loci in patients with UC or CD compared to healthy controls in terminal ileum and colon, respectively. It only contains genes which showed at least a two fold change in one comparison. Beside AQP8, genes like solute linked carrier 26A2 (SLC26A2), early growth response 1 (EGR1), mitogen-activated protein kinase kinase 2 (MAP2K2), protochlorophyllide oxidoreductase (POR), claudin 3 (CLDN3), glutamyl aminopeptidase (ENPEP), and eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) showed strong differential regulation. Among the identified genes AQP8 was of special interest because this gene displayed strong mRNA expression in normal ileum and colon. Furthermore, the Gene Chip analysis showed a specific expression pattern of the AQP8 gene in IBD patients. We noted that in the ileum the mRNA levels of AQP8 were severely reduced (-8.6 and -9.8 fold for CD and UC, respectively), whereas the expression of AQP8 was induced in the colon of both patient groups (4.3 and 3.0 fold for CD and UC, respectively).
| IBDcandidatelocus/Gene | Controlexpressionileum | FCCD | FCUC | Controlexpressioncolon | FCCD | FCUC |
| Chr. 16q11-12 | ||||||
| MMP2 | 267 | 1.5 | -1.1 | 220 | 1.5 | 3.7 |
| TM4SF11 | 194 | 1.3 | 1.0 | 151 | -2.1 | -1.7 |
| SIAH1 | 208 | 1.6 | 1.2 | 156 | 3.5 | 1.3 |
| MT1G | 3053 | -1.1 | 1.2 | 2356 | -1.7 | -2.0 |
| NETO2 | 49 | -1.6 | -1.7 | 61 | 2.5 | 1.4 |
| Chr. 12q13 | ||||||
| DGKA | 427 | -1.5 | 1.0 | 342 | -2.3 | -2.6 |
| MYO1A | 1008 | 1.1 | 1.1 | 811 | -3.5 | -4.3 |
| Chr. 5q32 | ||||||
| SPARC | 76 | 2.6 | 1.5 | 104 | 2.0 | 2.6 |
| TGFBI | 236 | 2.0 | 1.0 | 356 | 1.5 | 2.5 |
| CAMLG | 93 | 2.5 | 1.4 | 91 | 2.1 | 1.4 |
| TCF7 | 357 | -2.0 | -1.2 | 279 | -1.2 | -1.6 |
| SLC35A4 | 535 | -1.5 | -1.5 | 638 | -2.3 | -1.5 |
| SEC24A | 221 | 2.3 | 1.1 | 211 | 1.7 | 1.1 |
| SLC26A2 | 302 | 2.3 | -2.5 | 388 | 6.5 | -7.5 |
| EGR1 | 256 | 2.3 | 1.1 | 180 | 17.1 | 3.0 |
| PACAP | 150 | -2.8 | -1.7 | 138 | -1.6 | 2.0 |
| Chr. 19p13 | ||||||
| AES | 222 | -2.1 | -1,1 | 257 | -2.5 | -2.3 |
| MAP2K2 | 420 | -2.0 | -1,1 | 627 | -2.1 | -5.7 |
| Chr. 1p36 | ||||||
| CDC42 | 1428 | 1.1 | -1.2 | 1429 | 2.0 | -1.9 |
| MFN2 | 304 | -1,1 | 1.0 | 367 | -3.0 | -2.5 |
| C1QA | 336 | 1.1 | 1.1 | 373 | -1.6 | -2.5 |
| Chr. 16p12 | ||||||
| MIR16 | 201 | -1.7 | -1.1 | 277 | -3.0 | -1.2 |
| AQP8 | 575 | -8.6 | -9.8 | 438 | 4.3 | 3.0 |
| MT1G | 3053 | -1.1 | 1.2 | 2356 | -1.7 | -2.0 |
| LITAF | 500 | -1.2 | -1.2 | 670 | 1.9 | 2.1 |
| PRKCB1 | 157 | -2.0 | 1.4 | 95 | 1.1 | -1.2 |
| USP7 | 184 | 1.6 | -1.3 | 192 | 2.0 | -1.1 |
| Chr. 7q11 | ||||||
| WBSCR22 | 172 | -1.7 | -1.1 | 207 | -1.9 | -2.3 |
| POR | 213 | -2.1 | 1.0 | 172 | -2.3 | -4.9 |
| SEMA3C | 79 | 1.1 | 1.0 | 70 | 2.3 | 1.9 |
| CLDN3 | 386 | -1.4 | -1.2 | 618 | -2.6 | -6.5 |
| WBSCR20A | 323 | -1.6 | 1.0 | 281 | -2.0 | -1.7 |
| TMPIT | 928 | 1.1 | 1.1 | 893 | -2.3 | -2.3 |
| CLDN4 | 150 | 1.1 | 1.0 | 232 | -1.4 | -2.3 |
| CD36 | 153 | 2.8 | 1.3 | 127 | 2.3 | 1.1 |
| HSPB1 | 376 | -3.2 | -2.3 | 493 | -1.9 | -1.5 |
| Chr. 4q25 | ||||||
| ELOVL6 | 112 | 1.2 | -1.6 | 332 | 2.1 | 1.6 |
| ENPEP | 578 | 1.7 | 1.6 | 533 | 1.1 | -11.3 |
| T2BP | 322 | 1.2 | 1.1 | 224 | 2.8 | 1.4 |
| SYNPO2 | 242 | 2.3 | 1.3 | 160 | 2.5 | 1.1 |
| SEC24B | 105 | 2.0 | 1.6 | 161 | 1.6 | 1.1 |
| Chr. 3p21 | ||||||
| ACY1 | 744 | 1.1 | 1.2 | 485 | -2.1 | -1.5 |
| Chr. 3q27 | ||||||
| AP2M1 | 417 | -1.6 | 1.0 | 503 | -2.3 | -3.5 |
| TNIK | 143 | 2.1 | 1.1 | 135 | 1.4 | 1.0 |
| PRKCI | 68 | 2.1 | 1.2 | 104 | 2.0 | 1.1 |
| SI | 1313 | 1.3 | 1.1 | 963 | 1.4 | -2.0 |
| EIF4G1 | 292 | -2.5 | -1.6 | 319 | -3.7 | -6.1 |
| KLHL6 | 323 | -3.2 | 1.0 | 110 | 1.1 | 1.7 |
| FXR1 | 137 | 1.9 | 1.2 | 105 | 2.1 | 2.0 |
| Chr. 10q22 | ||||||
| MYST4 | 73 | 2.0 | 1.4 | 75 | 2.0 | 1.5 |
| RAI17 | 363 | 1.6 | 1.0 | 250 | 1.7 | 2.1 |
Apart from AQP8, only AQP1, AQP3, and AQP11 were expressed remarkably according to microarray data. For AQP0, AQP2, AQP4, AQP5, AQP6, AQP7, and AQP9 no transcripts could be detected, whereas AQP10 and AQP12 were not represented on the chip. The mRNA levels of AQP1, AQP3, and AQP11 were induced in the ileum of patients with UC or CD and reduced in the colon of patients with UC or CD (Table 5). Interestingly, this expression pattern was exactly inverse to the expression pattern of AQP8.
| Gene | Controlexpression ileum | FC CD | FC UC | Controlexpression colon | FC CD | FC UC |
| AQP1 | 181 | 1.7 | 1.7 | 165 | -2.0 | -1.6 |
| AQP3 | 473 | 1.9 | 1.5 | 352 | -1.4 | -3.2 |
| AQP8 | 575 | -8.6 | -9.8 | 438 | 4.3 | 3.0 |
| AQP11 | 510 | 1.2 | 1.0 | 409 | -1.9 | -2.0 |
In order to validate the microarray data, we have established a TaqMan real-time RT-PCR assay for AQP8 and examined the mRNA expression levels of AQP8 in each single control subject and patient used for pooling within the hybridization of the IBD microarrays. As obvious from Figure 1, the expression and dysregulation pattern observed in UC patients by microarray analysis could be confirmed by TaqMan real-time RT-PCR. For CD patients the microarray data were not reproducible by TaqMan real-time RT-PCR (data not shown).
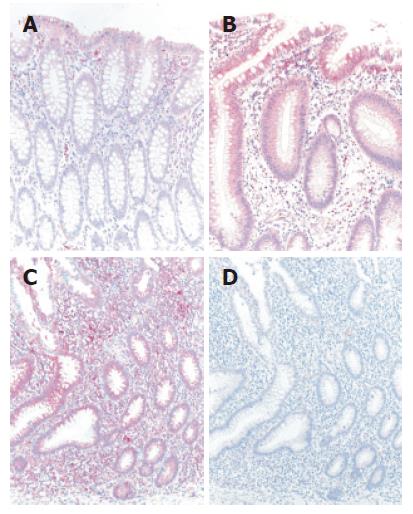
The differential mRNA expression observed in the colon of UC patients by microarray analysis and TaqMan real-time RT-PCR correlated with protein expression determined by immunohistochemistry (Figure 2). Normal colonic mucosa epithelial cells displayed very weak signals at the mucosal surface whereas a slight increase in staining intensity in an inactive area in a case with ulcerative colitis could be found. Both the intestinal epithelium as well as a subset of lamina propria mononuclear cells (MNCs) were positive for AQP8. Furthermore, in an actively inflamed colonic area in a case with ulcerative colitis a strong expression of AQP8 appeared. Intestinal epithelial cells as well as numerous MNCs within the inflamed lamina propria were immunostained.
Taken together, performing Affymetrix Gene Chip expression analysis we detected a significant upregulation of AQP8 mRNA levels in the colon of UC patients. This finding could be confirmed by both TaqMan RT-PCR and at protein level by immunohistochemistry.
Sequencing of the AQP8 gene and TaqMan allelic discrimination in patients with IBD and in healthy controls.
Since we identified the altered AQP8 mRNA levels in IBD and because AQP8 is located within the IBD locus 8, we speculated that genetic variations (mutations or polymorphisms) in the coding region or the regulatory region of AQP8 could predispose for the development of IBD. Therefore, we sequenced the AQP8 gene promoter region and its six exons in 100 CD patients, 100 UC patients, and in 50 healthy controls. As a genetic case-control association study TaqMan allelic discrimination in 220 patients with UC, in 181 patients with CD, and in 250 healthy controls was performed. Analyzing the sequencing results of the six exons and the promoter region 3 single nucleotide polymorphisms (SNPs) could be identified, each resulting in an amino acid change (Table 6). The SNP AQP8-A212T we found in the coding sequence of exon 5 causes an exchange of alanine into threonine in the AQP8 protein. The first SNP detected in exon 6 AQP8-A260P leads to an exchange of alanine into proline, the second one AQP8-R261Q to an exchange of arginin into glutamine. However, comparing the frequency of these SNPs in patients with UC or CD to the frequency in healthy controls no statistically significant difference could be found (Table 6).
| SNP | n VIC | % VIC | n FAM | % FAM | n both | % both | P-Value | χ2 | |
| AQP8-A212T | G | G | A | A | G/A | G/A | |||
| GCC -> ACC | UC | 210 | 95.5 | 0 | 0.0 | 9 | 4.1 | 0.57 | 0.33 |
| CD | 166 | 91.7 | 0 | 0.0 | 9 | 5.0 | 0.98 | 0.00 | |
| Co | 232 | 92.8 | 0 | 0.0 | 14 | 5.6 | |||
| AQP8-A260P | G | G | C | C | C/G | C/G | |||
| GCT -> CCT | UC | 94 | 42.7 | 33 | 15.0 | 78 | 35.5 | 0.26 | 2.66 |
| CD | 56 | 30.9 | 26 | 14.4 | 83 | 45.9 | 0.46 | 1.55 | |
| Co | 93 | 37.2 | 43 | 17.2 | 107 | 42.8 | |||
| APQ8-R261Q | G | G | A | A | G/A | G/A | |||
| CGG -> CAG | UC | 205 | 93.2 | 0 | 0.0 | 13 | 5.9 | 0.62 | 0.97 |
| CD | 159 | 87.8 | 1 | 0.6 | 9 | 5.0 | 0.97 | 0.07 | |
| Co | 214 | 85.6 | 1 | 0.4 | 13 | 5.2 |
CD and UC are chronic inflammatory disorders of the gastrointestinal tract, which are thought to result from the effect of environmental factors in genetically predisposed individuals. Mapping studies suggest a strong inherited component but a large number of putative susceptibility loci has complicated the identification of IBD genes. In our approach, we wanted to study the gene expression of mucosal cells from IBD patients in order to identify dysregulated target genes for potential therapy. Therefore, we decided to use DNA microarray analysis, which is a very powerful tool to perform large scale transcription profiling. So far, three groups have performed microarray analyses to examine gene expression in biopsies of IBD patients. One group analyzed tissue samples from inflamed mucosa of CD patients with a very limited cDNA array containing 96 genes and mainly identified aberrant expression of immune genes[16]. Two recent studies using the first[5] and the second[4] generation of Affymetrix arrays to screen samples from UC patients[5] or both CD and UC patients[4] obtained reproducible results. In our study, we used the Affymetrix Human Genome U133A and the U133B Gene Chips, to analyze pooled endoscopic tissue samples from non-inflamed areas of UC and CD patients. This array set captures the expression levels of 39 000 transcripts and variants, including more than 33 000 well-substantiated human genes. Pooling of RNA samples in microarray experiments is a very effective way to minimize biological variation of gene expression and reduces costs without a loss of precision[17]. Two important differences between our approach and that of the above mentioned groups are obvious. First, we have analyzed non-inflamed mucosa to avoid secondary inflammatory events. Consequently, this proceeding is suitable for revealing subclinical defects, because influx of inflammatory cell populations can profoundly change the transcriptional profile of the mucosa in IBD. Second, biopsies from two different gut sections, terminal ileum and colon, which have a markedly different pattern of gene expression were examined. In the context of our recently published study using the same microarray data[14], we could prove that our endoscopic biopsy samples mainly represented differentially regulated mRNA levels in epithelial cells. Combining the information from published linkage analysis and association studies with our Affymetrix microarray results we found a specific expression pattern of the AQP8 gene which is located on chromosome 16p12.1 and thus within the IBD locus 8. In the ileum of IBD patients we obtained severely reduced AQP8 mRNA levels whereas AQP8 expression was significantly induced in the colon of IBD patients. These findings could be confirmed both by TaqMan real-time RT-PCR and at protein level by immunohistochemistry for UC patients. Unfortunately, none of the commercially available antibodies used for Western Blotting did work in human biopsies. Therefore, no quantitative protein data can be shown. For CD patients the microarray data were not reproducible by TaqMan real-time RT-PCR. Our interpretation of this finding is that CD not only differs in clinical characteristics from UC but may also be caused by completely different underlying pathophysiologic mechanisms.
Although, about 10 liters of water are transported in the GI tract per day to perform the secretory and absorptive functions of the GI tract, the molecular basis of water secretion and absorption is hardly understood. Important progress in understanding water transport mechanisms in the GI tract was made by the recent identification of multiple epithelial AQP water channels indicating a key role in water secretion and absorption there[9,18-20]. In contrast to the colon where solutes are transported actively out of the crypt lumen across a relatively water-impermeable crypt barrier[21,22], rapid water movement in small intestine is generally believed to occur by a paracellular pathway. Furthermore, the small intestine has been proposed to be highly water permeable and to contain a highly convolved leaky epithelium. The cellular and subcellular distribution of AQP8 suggests physiological roles for this aquaporin in the absorption of water in the intestine. In addition, the cytoplasmic localization of AQP8 may also relate to the involvement in processes of intracellular osmoregulation[12]. Taking these facts into consideration, one could speculate that a dysregulation of the AQP8 mRNA expression could lead to disturbed processes in absorption of fluids in the GI tract as well as to disturbed processes of intracellular osmoregulation. An upregulation of AQP8 mRNA in the colon of UC patients may result in a dehydration and a higher viscosity of the adherent mucus layer which then affects the mucus adherence and finally disturbs the mucus barrier which protects the colonic mucosal cells against the attack of luminal bacteria by a continuous, hydrophobic and adherent mucus layer[23,24]. This assumption suits well to one of the current hypotheses concerning the pathophysiology of UC, namely that colonic commensal bacteria can attack the mucosa and contribute to the development of inflammation and ulceration in case of a disturbed mucosal barrier function[1,25-27]. The downregulation of AQP8 mRNA in the terminal ileum of UC patients may be a compensatory mechanism to avoid further dehydration of the mucus layer controlled by feedback mechanisms. Beside us, other authors like Ma and Verkman[28] can imagine that modulation of aquaporin function by novel pharmacological agents or gene delivery may alter the course of IBD or other GI disorders. Moreover, similar phenomena were described in other illnesses, e.g. cystic fibrosis (CF) where an alternative view of innate airway defense has emerged[29] which emphasizes a role for a chemical shield in protecting the lung against inhaled bacteria[30]. Especially as, aquaporins play an important role in lung physiology, it seems conceivable that CF lung disease reflects chronic depletion of the periciliary liquid layer volume which predicts adverse interactions between the mucus layer and the airway epithelial surface[31].
As one could suppose that genetic variations in the coding region or the regulatory region of AQP8 could be the reason for the dysregulation of AQP8 and thus predispose for IBD, we performed sequencing of the AQP8 gene and TaqMan allelic discrimination in IBD patients and healthy controls. In the end, no causal mutations or SNPs could be identified.
Though, from our data, the development of colonic inflammation in UC is associated with an altered expression of epithelial AQP8, the expression pattern of the AQP8 gene seems to be a secondary phenomenon due to another underlying cause. This theory is supported by our immunohistochemistry results, namely that we could find a further upregulation of AQP8 expression in case of inflammation. These changes may be the result of the inflammation but may nevertheless contribute to the pathophysiology of UC. Especially as recent studies suggest that fluid flux may play an important role in mucosal defense[32,33]. One could imagine that the upregulation of AQP8 in the colon of UC patients is due to signalling pathways which are not understood so far. Although the biopsies have been taken from non-inflamed areas cytokine effects are imaginable, since studies suggest a role for inflammatory mediators in the regulation of AQPs[34]. Our results are contrary to those of Hardin et al[35] who used biopsies from inflamed human colonic tissues. We suppose that the similar reductions in AQP expression they found in patients with UC, CD or infectious colitis is an unspecific phenomenon of inflammation. Even a change of the AQP8 expression dependent on the course of inflammation is conceivable. Maybe, at the very beginning of inflammation there is a downregulation of the AQP8 expression according to the findings of Hardin et al[35] which may lead to diarrhea and may be followed by a compensatory increase in expression.
In summary, we have been able to show that expression profiling of human tissue biopsies by DNA microarray technology is capable of identifying genes which may potentially play an important role in the pathogenesis of IBD. We could demonstrate a significant upregulation of the AQP8 gene in the colon of patients with UC which may be responsible for a disturbed mucus adherence and may consecutively allow an attack of luminal bacteria.
So far, several potential inflammatory bowel disease (IBD) susceptibility loci have been identified. In the absence of a priori candidate genes, experimental techniques such as microarray technology, seem to be an useful approach to genome-wide searches for IBD genes. Applying a mapping and arraying strategy, we obtained several genes of interest, among them, the aquaporin-8 (AQP8) gene, which is located within the IBD locus 8.
The article deals with the pathogenesis of ulcerative colitis. Related fields associated with the article are microarray technology, intestinal water transport mechanisms by aquaporins and the intestinal mucus barrier function.
Applying a mapping and arraying strategy, we obtained an interesting selection of differentially regulated genes located in IBD candidate loci in patients with ulcerative colitis (UC), among them, the AQP8 gene, which is located within the IBD locus 8. Both Gene Chip analysis and TaqMan RT-PCR showed severely reduced levels of AQP8 mRNA in the ileum of UC patients, whereas the expression of AQP8 was induced in the colon. Immunohistochemistry confirmed these findings. We concluded that an upregulation of AQP8 mRNA in the colon of UC patients may result in a dehydration and a higher viscosity of the adherent mucus layer which then affects the mucus adherence and finally disturbs the mucus barrier which protects the colonic mucosal cells against the attack of luminal bacteria by a continuous, hydrophobic and adherent mucus layer.
We can imagine that modulation of aquaporin function by novel pharmacological agents or gene delivery may alter the course of IBD or other GI disorders.
The aquaporins (AQPs) are a family of small (about 30 kDa) integral membrane proteins that function as water channels in animals, plants and bacteria. So far, 13 AQP homologues have been identified in mammals and an increasing number of disturbances have been found associated to the abnormal function of these proteins. Among mammalian aquaporins, two subgroups have been defined: “aquaporins” and “aquaglyceroporins”. The capability to transport glycerol as backbone molecule for triglycerides also links aquaporins to lipid metabolism, thus regional expression of AQPs may influence fatty acid metabolism in highly asorptive tissues such as the intestine.
The authors searched gene expression profiles of colonic and ileal mucosa in IBD and normal controls, and found that AQP8 expression is altered in IBD. They also evaluated mutations and polymorphisms in AQP8, and found no significant relation with IBD. They insist that change of AQP8 expression in IBD is due to altered water exchange. This report is very interesting, and revealing some probable pathogenetic mechanism of UC. The study is well designed.
S- Editor Liu Y L- Editor Rampone B E- Editor Chin GJ
| 1. | Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1493] [Cited by in RCA: 1484] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 2. | Rioux JD. Progress towards identifying inflammatory bowel disease susceptibility genes. Novartis Found Symp. 2004;263:3-11; discussion 11-16, 211-218. [PubMed] |
| 3. | Brant SR, Shugart YY. Inflammatory bowel disease gene hunting by linkage analysis: rationale, methodology, and present status of the field. Inflamm Bowel Dis. 2004;10:300-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 292] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 5. | Dieckgraefe BK, Stenson WF, Korzenik JR, Swanson PE, Harrington CA. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics. 2000;4:1-11. [PubMed] |
| 6. | Wayne ML, McIntyre LM. Combining mapping and arraying: An approach to candidate gene identification. Proc Natl Acad Sci USA. 2002;99:14903-14906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 180] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 7. | Viggiano L, Rocchi M, Svelto M, Calamita G. Assignment of the aquaporin-8 water channel gene (AQP8) to human chromosome 16p12. Cytogenet Cell Genet. 1999;84:208-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 8. | Huang P, Lazarowski ER, Tarran R, Milgram SL, Boucher RC, Stutts MJ. Compartmentalized autocrine signaling to cystic fibrosis transmembrane conductance regulator at the apical membrane of airway epithelial cells. Proc Natl Acad Sci USA. 2001;98:14120-14125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 166] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 9. | King LS, Yasui M, Agre P. Aquaporins in health and disease. Mol Med Today. 2000;6:60-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 10. | Echevarría M, Ilundáin AA. Aquaporins. J Physiol Biochem. 1998;54:107-118. [PubMed] |
| 11. | Krane CM, Fortner CN, Hand AR, McGraw DW, Lorenz JN, Wert SE, Towne JE, Paul RJ, Whitsett JA, Menon AG. Aquaporin 5-deficient mouse lungs are hyperresponsive to cholinergic stimulation. Proc Natl Acad Sci USA. 2001;98:14114-14119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 90] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Calamita G, Mazzone A, Bizzoca A, Cavalier A, Cassano G, Thomas D, Svelto M. Expression and immunolocalization of the aquaporin-8 water channel in rat gastrointestinal tract. Eur J Cell Biol. 2001;80:711-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Fischer H, Stenling R, Rubio C, Lindblom A. Differential expression of aquaporin 8 in human colonic epithelial cells and colorectal tumors. BMC Physiol. 2001;1:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Langmann T, Moehle C, Mauerer R, Scharl M, Liebisch G, Zahn A, Stremmel W, Schmitz G. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 259] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 15. | Langmann T, Mauerer R, Zahn A, Moehle C, Probst M, Stremmel W, Schmitz G. Real-time reverse transcription-PCR expression profiling of the complete human ATP-binding cassette transporter superfamily in various tissues. Clin Chem. 2003;49:230-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 201] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 16. | Heller RA, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley DE, Davis RW. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA. 1997;94:2150-2155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 481] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 17. | Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 160] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Ishibashi K, Sasaki S, Fushimi K, Uchida S, Kuwahara M, Saito H, Furukawa T, Nakajima K, Yamaguchi Y, Gojobori T. Molecular cloning and expression of a member of the aquaporin family with permeability to glycerol and urea in addition to water expressed at the basolateral membrane of kidney collecting duct cells. Proc Natl Acad Sci USA. 1994;91:6269-6273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 436] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 19. | Jung JS, Bhat RV, Preston GM, Guggino WB, Baraban JM, Agre P. Molecular characterization of an aquaporin cDNA from brain: candidate osmoreceptor and regulator of water balance. Proc Natl Acad Sci USA. 1994;91:13052-13056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 523] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 20. | Koyama Y, Yamamoto T, Kondo D, Funaki H, Yaoita E, Kawasaki K, Sato N, Hatakeyama K, Kihara I. Molecular cloning of a new aquaporin from rat pancreas and liver. J Biol Chem. 1997;272:30329-30333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 130] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Naftalin RJ, Pedley KC. Video enhanced imaging of the fluorescent Na+ probe SBFI indicates that colonic crypts absorb fluid by generating a hypertonic interstitial fluid. FEBS Lett. 1990;260:187-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Naftalin RJ. The dehydrating function of the descending colon in relationship to crypt function. Physiol Res. 1994;43:65-73. [PubMed] |
| 23. | Lichtenberger LM. The hydrophobic barrier properties of gastrointestinal mucus. Annu Rev Physiol. 1995;57:565-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 219] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 24. | Wallace JL, Granger DN. The cellular and molecular basis of gastric mucosal defense. FASEB J. 1996;10:731-740. [PubMed] |
| 25. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2747] [Article Influence: 119.4] [Reference Citation Analysis (2)] |
| 26. | Rath HC, Schultz M, Freitag R, Dieleman LA, Li F, Linde HJ, Schölmerich J, Sartor RB. Different subsets of enteric bacteria induce and perpetuate experimental colitis in rats and mice. Infect Immun. 2001;69:2277-2285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 241] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 27. | Veltkamp C, Tonkonogy SL, De Jong YP, Albright C, Grenther WB, Balish E, Terhorst C, Sartor RB. Continuous stimulation by normal luminal bacteria is essential for the development and perpetuation of colitis in Tg(epsilon26) mice. Gastroenterology. 2001;120:900-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Ma T, Verkman AS. Aquaporin water channels in gastrointestinal physiology. J Physiol. 1999;517:317-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 210] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 29. | Smith JJ, Travis SM, Greenberg EP, Welsh MJ. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 741] [Cited by in RCA: 711] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 30. | Bals R, Weiner DJ, Wilson JM. The innate immune system in cystic fibrosis lung disease. J Clin Invest. 1999;103:303-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 135] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 31. | Knowles MR, Boucher RC. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest. 2002;109:571-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 780] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 32. | Asfaha S, MacNaughton WK, Appleyard CB, Chadee K, Wallace JL. Persistent epithelial dysfunction and bacterial translocation after resolution of intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2001;281:G635-G644. [PubMed] |
| 33. | Morris GP, Fallone CA, Pringle GC, MacNaughton WK. Gastric cytoprotection is secondary to increased mucosal fluid secretion: a study of six cytoprotective agents in the rat. J Clin Gastroenterol. 1998;27 Suppl 1:S53-S63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Steinfeld SD, Appelboom T, Delporte C. Treatment with infliximab restores normal aquaporin 5 distribution in minor salivary glands of patients with Sjögren's syndrome. Arthritis Rheum. 2002;46:2249-2251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | Hardin JA, Wallace LE, Wong JF, O'Loughlin EV, Urbanski SJ, Gall DG, MacNaughton WK, Beck PL. Aquaporin expression is downregulated in a murine model of colitis and in patients with ulcerative colitis, Crohn's disease and infectious colitis. Cell Tissue Res. 2004;318:313-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 87] [Article Influence: 4.1] [Reference Citation Analysis (0)] |