Published online Feb 28, 2006. doi: 10.3748/wjg.v12.i8.1192
Revised: October 25, 2005
Accepted: November 10, 2005
Published online: February 28, 2006
AIM: To screen a suspected Hungarian HNPCC family to find specific mutations and to evaluate their effect on the presentation of the disease.
METHODS: The family was identified by applying the Amsterdam and Bethesda Criteria. Immunohistochemistry was performed, and DNA samples isolated from tumor tissue were evaluated for microsatellite instability. The identification of possible mutations was carried out by sequencing the hMLH1 and hMSH2 genes.
RESULTS: Two different mutations were observed in the index patient and in his family members. The first mutation was located in exon 7, codon 422 of hMSH2, and caused a change from Glu to STOP codon. No other report of such a mutation has been published, as far as we could find in the international databases. The second mutation was found in exon 3 codon 127 of the hMSH2 gene, resulting in Asp→Ser substitution. The second mutation was already published, as a non-pathogenic allelic variation.
CONCLUSION: The pedigree analysis suggested that the newly detected nonsense mutation in exon 7 of the hMSH2 gene might be responsible for the development of colon cancers. In family members where the exon 7 mutation is not coupled with this missense mutation, colon cancer appears after the age of 40. The association of these two mutations seems to decrease the age of manifestation of the disease into the early thirties.
- Citation: Tanyi M, Olasz J, Lukács G, Csuka O, Tóth L, Szentirmay Z, Ress Z, Barta Z, Tanyi JL, Damjanovich L. Pedigree and genetic analysis of a novel mutation carrier patient suffering from hereditary nonpolyposis colorectal cancer. World J Gastroenterol 2006; 12(8): 1192-1197
- URL: https://www.wjgnet.com/1007-9327/full/v12/i8/1192.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i8.1192
According to published data, about 15-20 % of colorectal carcinomas (CRC) follow a familial pattern. Familial adenomatous polyposis coli syndrome (FAP) and its subtypes are responsible for only about 1 % of all CRCs, while hereditary nonpolyposis colon cancer (HNPCC) accounts for approximately 3-8 % of cases[1-3]. The characteristic presentation of HNPCC is frequently right-sided localization, the presence of synchronous and metachronous CRCs, and its association with other HNPCC-related extracolonic tumors, including gastric, endometrial, and urinary and biliary tract cancers in afflicted families[2,3]. Compared to sporadic colorectal carcinomas, HNPCC has an earlier age of onset, Crohn’s disease-like lymphocytic infiltration in tumor tissues, increased mucin production, and a lower degree of histological differentiation[3-5]. The classic adenoma-carcinoma sequence can be observed in HNPCC patients as well, but the conversion of polyps to malignant proliferation is accelerated, taking only 1-3 years, as opposed to sporadic adenomas, where this can take as long as 10-15 years[1]. The number of polyps in the colon is not so high as in cases of FAP. The genetic background of HNPCC has been identified in the germline defect of DNA mismatch repair genes (MMR). The syndrome is inherited in an autosomal dominant fashion, with an estimated penetrance of 80%[1-3,6]. At present, seven MMR genes are known (hMLH1, hMLH3, hMSH2, hMSH3, hMSH6, hPMS1, hPMS2); however, the influence of a specific mutation on the pathogenesis of the disease shows great diversity[7-10]. In about 90-95% of the cases, germline mutations of the hMLH1 or hMSH2 genes can be demonstrated, while only a low percentage is caused by the mutations of hMSH6, hPMS1 and hPMS2[1]. The detection of afflicted families must be initiated by taking a thorough family history, embracing at least three generations, applying the widely accepted Amsterdam and Bethesda Criteria[6]. If the diagnosis of HNPCC is further supported by the immunohistology of the MMR proteins, the microsatellite status must be determined. The hallmark of mismatch repair deficiency is microsatellite instability (MSI), variability of the lengths of short repetitive DNA stretches scattered in the genome (microsatellites) compared to normal tissue DNA[11-14]. Finally, sequencing of the MMR genes can accurately identify mutations and makes screening of other family members possible[6]. It is emphasized in the literature that only around 40% of the families identified by the Amsterdam Criteria will actually present detectable mutations. This fact influences both therapeutic plans and the required long-term follow-up[3,13,15,16].
In this study, our goal was to perform the genetic analysis of a young male patient and the members of his family who had been selected by the Amsterdam Criteria, and to identify the genetic alterations in the MMR genes hMLH1 and hMSH2. We also attempted to establish a correlation between the occurrence of mutations and disease pattern.
A 32-year-old male patient (index person) with recurrent hematochesia was received at an outpatient clinic in North-Eastern Hungary. Colonoscopy was performed and a friable, bleeding lesion was found at the lienal flexure of the large bowel with circular narrowing of the lumen. The histopathological report of the biopsy showed a mucinous adenocarcinoma infiltrating the muscularis propria, pT2, Grade 2. Ultrasound and computer tomography ruled out evidence of liver metastases, enlarged lymph nodes in the mesocolon or spread to neighboring organs. Since the disease was localized, a left hemicolectomy was performed with an end-to-end transverso-rectostomy. The family and the index person were evaluated using the Amsterdam and Bethesda Criteria[2]. Based on the family history, the possible diagnosis of HNPCC was suspected and, following discussions about the disease, the patient and his family agreed to further genetic evaluation.
Routine 5 μm, paraffin-embedded tissue sections were dewaxed, rehydrated, and treated in a microwave oven in 10 mmol/L citrate buffer (pH 6.4) for 20 min, in order to retrieve antigenecity. Unspecific protein binding was blocked with 1% bovine serum albumin containing PBS for 30 min at 37 °C, then slides were incubated overnight with the primary antibodies (clone 2 MSH01, mouse monoclonal anti-MSH2, 1:100 and clone M074581 mouse monoclonal anti-MLH1, Labvision Corp., Fermont, CA, USA), respectively. Primary antibodies were detected by a biotin-streptavidine detection kit (LSAB, Dako, Carpinteria, CA, USA) using VIP chromogen. Negative controls were stained with the omission of the primary antibodies.
DNA was extracted from a paraffin-embedded cancerous tissue sample of the index patient following deparaffinization and proteinase K digestion, according to the protocol of the High Pure PCR Template Purification kit (Roche Diagnostics GmbH, Mannheim, Germany). DNA samples of the patient and family members were also isolated from whole blood using the Wizard DNA Purification System (Promega Corporation, Madison WI, USA) according to Promega’s Technical Manual (TM050).
DNA samples isolated from the tumor tissue were tested for microsatellite instability. Two mononucleotide markers (BAT25 and BAT26) were evaluated using the technique described by Dietmaier et al[17]. A multiplex PCR was performed using sequence specific hybridization probes (HyProbes) labeled with LightCycler dyes, LCRed640 and LCRed705. Amplification of microsatellites by LightCycler (Roche Diagnostics GmbH, Germany) was followed by melting point analysis to display alterations in the length of repetitive sequences.
All exons of hMLH1 and hMSH2 genes were analyzed in the blood sample of the index patient. Primers and PCR conditions were used as previously described[18]. Primers are available upon request. After denaturation or heteroduplex formation, the PCR products were loaded to electrophoresis on MDE gel (Cambrex Bio Science Rockland Inc., Rockland, MA, USA) according to manufacturer’s instructions and visualized by silver staining.
PCR products showing altered migration patterns on MDE gel were purified and sequenced in both directions. Sequencing reactions were performed using a BigDye thermocycler sequencing kit v.3.1 (Applied Biosystems, Foster City, CA, USA). The semi-automated fluorescence analysis was performed with an ABI-PRISM 310 Genetic Analyzer (Perkin Elmer, Boston, MA, USA).
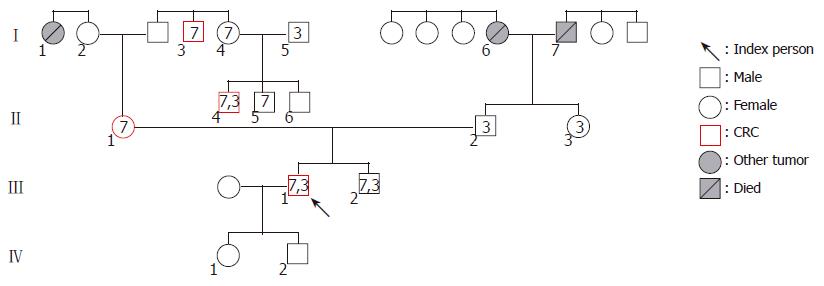
After a comprehensive evaluation of three generations, the index person’s family was found to fulfill several points of the Amsterdam and Bethesda Criteria. In the maternal line of the pedigree, seven persons carry the exon 7 mutation. In this line, four cases of colorectal cancer and one case of breast cancer were recognized spanning three generations among first degree relatives. Analyzing the history of the paternal line, pulmonary carcinoma and gastric carcinoma (one case each) were detected (Figure 1 and Table 1). We found two generations (I./5 and II./2) in the maternal line, where exon 3 mutations join from two different families. In both cases of mutation associations, the tumor manifestation could be observed at a younger age than in the other family members, who carried the exon 7 mutation only. Numbers in icons mean the location of hMSH2 exon mutations. Numbers below icons identify the investigated family members. The younger brother of the index person (III./2.) was 28 years old at the time of the investigation and was disease-free.
| Classification | Degree of relationship | 7 exonmutation | 3 exonmutation | Type of the tumor,age at tumor detection | Age at investigation |
| I/1 | Sister of the mother of the index person’s mother | Breast cancer, 61 yr | Died | ||
| I/2 | Mother of the index person’s mother | 76 yr | |||
| I/3 | Brother of the father of the index person’s mother | + | CRC, 56 yr | 68 yr | |
| I/4 | Sister of the father of the index person’s mother | + | 61 yr | ||
| I/5 | Husband of the person No I/4 | + | 64 yr | ||
| I/6 | Mother of the index person’s father | Gastric cancer, 55 yr | Died | ||
| I/7 | Father of the index person’s father | Lung cancer, 75 yr | Died | ||
| II/1 | Index person’s mother | + | CRC, 43 yr | 54 yr | |
| II/2 | Index person’s father | + | 58 yr | ||
| II/3 | Sister of index person’s father | + | 48 yr | ||
| II/4 | The middle son of person No I/4 | + | + | CRC, 34 yr | 36 yr |
| II/5 | The youngest son of person No I/4 | + | 32 yr | ||
| II/6 | The oldest son of person No I/4 | 39 yr | |||
| III/1 | Index person | + | + | CRC, 31 yr | 33 yr |
| III/2 | Index person’s younger brother | + | + | 28 yr | |
| IV/1 | Older son of the index person | 3 yr | |||
| IV/2 | Younger son of the index person | 1 yr |
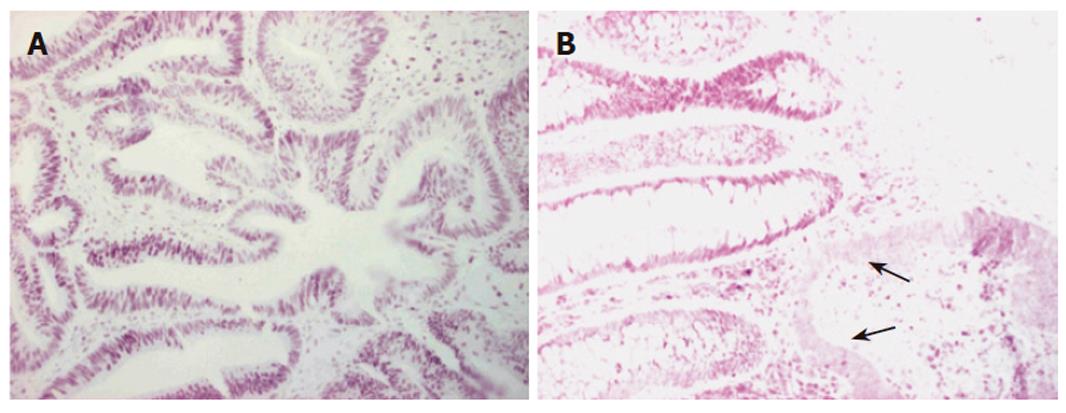
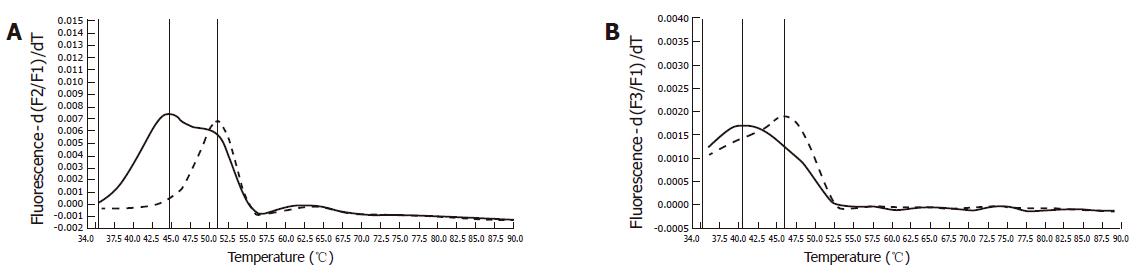
Immunohistochemistry of the index person’s tumor tissue showed a complete loss of hMSH2 reactivity. The hMLH1 staining was retained in the nuclei of the tumor cells (Figure 2). Furthermore, this tumor tissue was also proved to be MSI-High, as was demonstrated by melting point analysis of both mononucleotide markers, BAT25 and BAT 26 (Figure 3).
Sequencing of all the exons of hMSH2 and hMLH1 genes from the same tumor tissue showed two mutations of the hMSH2 gene. We were not able to demonstrate any alterations in the hMLH1 gene. The first mutation was a nonsense mutation, located in exon 7, codon 422 causing a glutamine→STOP codon change (Table 2). The second one was a missense mutation, located in exon 3, codon 127 resulting in substitution of Asp for Ser.
| Gene | Exon | Codon | Mutation | Type |
| hMSH2 | 7 | 422 | gaa→taa Glu→STOP | Nonsense |
| hMSH2 | 3 | 127 | aat→agt Asn→Ser | Missense |
Targeted sequencing of these two identified mutations was performed on DNA samples isolated from the peripheral blood leukocytes of fourteen other family members. On the paternal side, the missense mutation of exon 3 was identified in the index patient’s younger brother and his father, as well as in the sister of the father. Interestingly, the exon 3 mutation was also found in two members of the maternal line. The nonsense mutation of exon 7 was found in seven members of the maternal side and four of these subjects have been diagnosed as CRC (Table 1).
A characteristic feature of HNPCC is a tendency to develop metachronous tumors in affected individuals. In a 15-year follow-up study, Jarvinen and coworkers demonstrated convincingly that recurrence of CRC can be eliminated effectively by regular interval colonoscopies and polypectomies. Out of the 133 individuals who underwent regular interval follow-up examinations, 18% developed colon cancer, but no patients died. Whereas, among the 119 patients who deferred colonoscopy at three-year intervals, 41% were diagnosed as CRC and 9 deaths occurred[19]. Since the correct identification of HNPCC families is still an unsolved issue, considerable effort is dedicated to the task of uncovering further details of the disease. The most widely applied initial screening tools are the Amsterdam and Bethesda Criteria[2,3,6]. Unfortunately, neither of them is accurate enough to identify all HNPCC families with complete certainty. The most reliable, currently available technique is the demonstration of mutations in the MMR genes of suspected index persons, followed by a comprehensive search for the specific mutation in their family members. A promising way to reach an effective screening tool is to find and describe the nature and frequency of population specific mutations and to test these first in patients from a particular geographic location. Currently, more than 300 different MMR gene mutations have been published from different parts of the world. Some of these are recurrent, and have been described as founder mutations in particular populations[20-24] .
Our group, applying the previously suggested protocol, initiated an investigation in Hungary. A 31-year-old male patient was selected by the Bethesda criteria and genetic testing identified him as having HNPCC. A double mutation of the hMSH2 gene was found and subsequently compared with the available international databases (The Human Gene Mutation Database, Cardiff, International Society for Gastrointestinal Hereditary Tumors). The nonsense mutation of exon 7 has not been published before; therefore, it may potentially be characteristic in Hungarian families. The genetic error clearly appears to be pathogenic, since all the family members with CRC carry this mutation. The newly identified mutation causes development of a STOP codon, leading to a non-functional, truncated protein, which could not be detected by immunohistochemistry. The human MSH2 protein interacts with MSH6 forming the complex called hMutSα and with hMSH3 protein forming the complex named hMutSβ. Each subunit of these heterodimers is formed by five flexible domains which are defined as I. mismatch bindig, II. connector, III. core, IV. DNA clamp, and V. ATPase domains. The mismatch binding and DNA clamp domains act in binding to mismatched DNA whereas the core and connector domains constitute the backbone of each subunit. The latter two transmit allosteric information of bound DNA co-factor to the ATPase domain. Our detected mutation at codon 422 causing Glutamine→STOP change results in the loss of more than half of the expressed MSH2 protein ( truncated by 513 aminoacids from 934), involving core, DNA clamp and ATPase domains. This nonfunctional truncated protein cannot take a part in forming normal heterodimers and disintegrated quite soon after expression. To the best of the authors’ knowledge there was only one publication in Hungary of a novel hMSH2 germline mutation causing truncated protein expression. Czakó et al. investigated a 62 year old female patient who suffered from colon cancer at the age of 46, rectum cancer at the age of 60, endometrial cancer at the age of 56 years, basosquamous and squamous cell cancer of the face at the ages of 53, 54, 62, and 58 years respectively. This patient’s family fulfilled the Amsterdam criteria. The DNA sequencing analysis revealed a mutation as a single nucleotide change at codon 2292 in exon 14 of the hMSH2 gene. This guanin to adenin change altered the 764 amino acid, the tryptophan to STOP codon. Thus the hMSH2 protein was truncated by 171 aminoacids[25].
The missense mutation of exon 3 has already been described by several authors as a single amino acid change, and represents a non-pathogenic polymorphism of the gene. Samowitz et al published data showing 3 cases of this polymorphism in 1066 screened subjects. One of the 3 affected carriers was also found to have another missense mutation (exon 6, codon 328, Ala→Pro change, of hMSH2 gene), and in another carrier it was coupled with a pathogenic frameshift mutation of the hMLH1 gene (exon 16, codon 626).[26] De la Chapelle et al published another interesting double mutation, where this polymorphism was associated with an MSH2, exon 6, codon 333 mutation (International Society for Gastrointestinal Hereditary Tumors). The allele frequency of this polymorphism, according to the database of The National Institute of Environmental Health Sciences Genome Project, is 0.02, which would lead to as much as a 4% prevalence of heterozygous carriers in the general population.
Both mutations identified in this study are single nucleotide changes of the hMSH2 gene. Large genomic deletions and/or rearrangements, on the other hand, can account for 36 % of all hMSH2 mutations according to the Dutch HNPCC mutation analysis study.[27] Unlike the hMSH2 mutations, the majority of hMLH1 mutations are single nucleotide changes, but interestingly, a 3.5 kb deletion encompassing exon 16 of hMLH1 has been observed in Finland, as a founder mutation[23].
The presence of the exon 7 mutation alone resulted in a tumor manifestation at an age of 43 and 56 years (Table 1). When the missense and nonsense mutations were inherited together, the age of manifestation shifted to a younger age of 31 and 34 years.
It is well known that the penetrance of the MMR genes is less then 100%, which might explain why two of the older family members carrying the pathogenic mutation have not acquired the disease yet. Young age may answer the same question in the case of the younger brother of the index person, who was 28 years old at the time of the investigation (Table 1). On the other hand, all these family members must be considered “high-risk” patients, and be placed under close regular interval surveillance. To date, three family members carrying only the exon 3 mutation are disease-free. Three other members of the family, who suffered from breast, gastric and pulmonary cancers, respectively, could not be investigated because of their deaths before the initiation of the study.
Both the paternal and maternal lines of the evaluated family in this study came from the same small village in Hungary, which may explain the high prevalence of the exon 3 mutation of MSH2 in the tested individuals. It will be interesting to see whether the exon 7 mutation is truly characteristic for Hungarian families, or whether this mutation can be found in different populations elsewhere.
We are grateful to all the family members for their full cooperation during this study.
S- Editor Guo SY L- Editor Zhang JZ E- Editor Liu WF
| 1. | Souza RF. A molecular rationale for the how, when and why of colorectal cancer screening. Aliment Pharmacol Ther. 2001;15:451-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 2. | Scaife CL, Rodriguez-Bigas MA. Lynch syndrome: implications for the surgeon. Clin Colorectal Cancer. 2003;3:92-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | Steinkamp RC. Mortality rates from carcinoma of the uterine cervix in Arkansas: 1950-1969. J Ark Med Soc. 1975;71:312-313. [PubMed] |
| 4. | Jass JR. Role of the pathologist in the diagnosis of hereditary non-polyposis colorectal cancer. Dis Markers. 2004;20:215-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 5. | Pucciarelli S, Agostini M, Viel A, Bertorelle R, Russo V, Toppan P, Lise M. Early-age-at-onset colorectal cancer and microsatellite instability as markers of hereditary nonpolyposis colorectal cancer. Dis Colon Rectum. 2003;46:305-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Debniak T, Kurzawski G, Gorski B, Kladny J, Domagala W, Lubinski J. Value of pedigree/clinical data, immunohistochemistry and microsatellite instability analyses in reducing the cost of determining hMLH1 and hMSH2 gene mutations in patients with colorectal cancer. Eur J Cancer. 2000;36:49-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920-3923. [PubMed] |
| 8. | Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA, Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215-1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1588] [Cited by in RCA: 1485] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 9. | Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1019] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 10. | Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1335] [Cited by in RCA: 1265] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 11. | Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455-3460. [PubMed] |
| 12. | Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 97] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Calistri D, Presciuttini S, Buonsanti G, Radice P, Gazzoli I, Pensotti V, Sala P, Eboli M, Andreola S, Russo A. Microsatellite instability in colorectal-cancer patients with suspected genetic predisposition. Int J Cancer. 2000;89:87-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 14. | Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545-4549. [PubMed] |
| 15. | González-Aguilera JJ, Nejda N, Fernández FJ, Medina V, González-Hermoso F, Barrios Y, Moreno Azcoita M, Fernández-Peralta AM. Genetic alterations and MSI status in primary, synchronous, and metachronous tumors in a family with hereditary nonpolyposis colorectal cancer (HNPCC). Am J Clin Oncol. 2003;26:386-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Pawlik TM, Raut CP, Rodriguez-Bigas MA. Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers. 2004;20:199-206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 102] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 17. | Dietmaier W, Hofstädter F. Detection of microsatellite instability by real time PCR and hybridization probe melting point analysis. Lab Invest. 2001;81:1453-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Beck NE, Tomlinson IP, Homfray T, Frayling I, Hodgson SV, Harocopos C, Bodmer WF. Use of SSCP analysis to identify germline mutations in HNPCC families fulfilling the Amsterdam criteria. Hum Genet. 1997;99:219-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Järvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomäki P, De La Chapelle A, Mecklin JP. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 906] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 20. | Peltomäki P, Gao X, Mecklin JP. Genotype and phenotype in hereditary nonpolyposis colon cancer: a study of families with different vs. shared predisposing mutations. Fam Cancer. 2001;1:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Froggatt NJ, Green J, Brassett C, Evans DG, Bishop DT, Kolodner R, Maher ER. A common MSH2 mutation in English and North American HNPCC families: origin, phenotypic expression, and sex specific differences in colorectal cancer. J Med Genet. 1999;36:97-102. [PubMed] |
| 22. | Jäger AC, Bisgaard ML, Myrhøj T, Bernstein I, Rehfeld JF, Nielsen FC. Reduced frequency of extracolonic cancers in hereditary nonpolyposis colorectal cancer families with monoallelic hMLH1 expression. Am J Hum Genet. 1997;61:129-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Nyström-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Järvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med. 1995;1:1203-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 206] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 24. | Nyström-Lahti M, Wu Y, Moisio AL, Hofstra RM, Osinga J, Mecklin JP, Järvinen HJ, Leisti J, Buys CH, de la Chapelle A. DNA mismatch repair gene mutations in 55 kindreds with verified or putative hereditary non-polyposis colorectal cancer. Hum Mol Genet. 1996;5:763-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 150] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Czakó L, Tiszlavicz L, Takács R, Baradnay G, Lonovics J, Cserni G, Závodná K, Bartosova Z. [The first molecular analysis of a Hungarian HNPCC family: a novel MSH2 germline mutation]. Orv Hetil. 2005;146:1009-1016. [PubMed] |
| 26. | Samowitz WS, Curtin K, Lin HH, Robertson MA, Schaffer D, Nichols M, Gruenthal K, Leppert MF, Slattery ML. The colon cancer burden of genetically defined hereditary nonpolyposis colon cancer. Gastroenterology. 2001;121:830-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 27. | Wijnen J, van der Klift H, Vasen H, Khan PM, Menko F, Tops C, Meijers Heijboer H, Lindhout D, Møller P, Fodde R. MSH2 genomic deletions are a frequent cause of HNPCC. Nat Genet. 1998;20:326-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 154] [Article Influence: 5.7] [Reference Citation Analysis (0)] |