MATERIALS AND METHODS
Chemicals and antibodies
RPMI-1640 and sodium pyruvate were purchased from Biochrom, Berlin, Germany. Penicillin, streptomycin and fetal bovine serum (FBS) were purchased from Gibco, Germany. Primary antibodies such as anti-p53 and anticaspase-3 were purchased from Novocastra Laboratories Ltd, Newcastle, UK and Transduction Laboratories, Lexington, UK, respectively.
Drug preparation
T. arjuna bark was coarsely powdered and soaked in 95% ethanol and kept for 10 d at room temperature for maceration, filtered, concentrated and dried in a vacuum evaporator. Ethanolic extract of T. arjuna was dissolved in 1% DMSO [final concentration of the DMSO did not exceed 1% (v/v) and did not affect the cell proliferation] prepared in serum free RPMI medium and filtered by 0.3 mm syringe filter and stored.
HepG2 cell maintenance
Human hepatoma cell line (HepG2) was obtained from National Center for Cell Science, Department of Biotechnology, Pune, India. Cells were grown as monolayers in RPMI-1640 medium, supplemented with 10% (v/v) heat-inactivated FBS, antibiotics (penicillin 100U/mL, streptomycin 10 μg/mL) and 1mmol/L sodium pyruvate under standard conditions (37 oC) in a controlled humidified atmosphere containing 50 mL/L CO2.
Cytotoxicity
Trypan blue exclusion method: The cytotoxicity of the ethanolic extract of T. arjuna was assessed by cell viability using trypan blue exclusion method. For the determination of cell viability, cells were plated at the density of 5×104 cells/well and cultured for 48 h. The medium was replaced with serum-free medium [RPMI-1640 medium, supplemented with antibiotics (penicillin100 U/mL, streptomycin10 μg/mL, 1 mmol/L sodium pyruvate) and the cells were treated with various concentrations of T. arjuna (20, 40, 60, 80 and 100 mg/L) for 48 h and incubated with 1% DMSO as solvent control.
Lactate dehydrogenase leakage assay: Lactate dehydrogenase (LDH) leakage assay was performed by the method of Grivell and Berry [7]. One hundred microliters of conditioned media of control and T. arjuna-treated HepG2 cells was added to 1-mL cuvette containing 0.9mL of a reaction mixture to yield a final concentration of 1mmol/L pyruvate, 0.15 mmol/L NADH and 104 mmol/L phosphate buffer, pH7.4. After thoroughly mixed, the absorbance of the solution was measured at 340 nm for 45 s. LDH activity was expressed as μ moles of NADH used per minute per well.
Apoptosis
Light microscopic studies: HepG2 cells were grown in 35 mm sterile petri plates and treated with T. arjuna extract at the concentrations of 60 and 100 μg/L for 48 h. The cells were then fixed for 5 min with 10% methanol/phosphate buffer saline (PBS) and morphological changes were observed under inverted microscope (Nikon, Japan).
Fluorescent microscopic studies: Apoptotic morphology was studied by staining the cells with a combination of the fluorescent DNA-binding dye. After treatment with T. arjuna (60 and 100 mg/L for 48 h), cells were collected, washed and suspended in PBS. After stained with the equal mixture of acridine orange and ethidium bromide (each dye was dissolved in 100 μg/mL of PBS), the cells were examined[8]. The differential uptake of these two dyes allowed the identification of viable and non-viable cells under fluorescent microscope and the results were recorded.
DNA fragmentation: DNA fragmentation was followed by the method of Chen et al [9]. The HepG2 cells were plated in a 60 mm culture dish at a density of 5×105 cells and treated with T. arjuna extract (60 and 100 mg/L) for 48 h. The cells attached at the bottom were scraped off and collected together with unattached cells by centrifuging at 1 500 r/min for 5 min at 4 oC. DNA was prepared from the pelleted cells. Briefly, the cells were lysed with lysis buffer and extracted with 2mL of phenol (neutralized with TE buffer, pH 7.5) followed by extraction with 1mL of chloroform and isoamyl alcohol in the ratio of 24:1. The aqueous supernatant was precipitated with 2.5 volume of ice-cold ethanol with 10% volume of sodium acetate at –20 oC overnight. After centrifugation at 13 000 r/min for 10 min, the pellets were air-dried and resuspended with 50 μL of Tris–EDTA (TE) buffer. Equal quantities of DNA were electrophoresed in 1.8% agarose gel containing 0.5 mg/L of ethidium bromide. After electrophoresis, the gel was photographed under UV light.
Western blotting: HepG2 cells (1×106/mL) were treated with T. arjuna extract at the concentration of 60 and 100 μg/mL for 48 h at 37 oC. Cells were lysed with 10 μL of lysis buffer (150mmol/L Tris-HCl, 2 mmol/L N-ethylmaleimide, 2 mmol/L phenylmethyl sulfonyl fluoride, 4% sodium dodecylsulfate, 4% dithiothreitol, 20% glycerol, 0.01% bromophenol blue, 2 mol/L urea and 10mmol/L Na-EDTA) (pH 6.8). Cells were then sonicated three times for 10 s. Cell lysates were centrifuged at 15 000 r/min for 20 min at 4 oC. The protein levels were quantified using Biorad protein assay kit, USA. Equivalent amounts of proteins (60μg) were resolved by SDS-PAGE and transferred onto a nitrocellulose membrane overnight at 25 mA. Membranes were blocked with blocking solution (2% BSA in PBS) for 1 h at 37 oC followed by incubation with primary antibodies (anti-p53 1:100; anticaspase-3 1:1 000) for 1 h under gentle agitation. The blots were washed three times and incubated with horse raddish peroxidase-conjugated antimouse IgG antibody for 1 h at 37 oC. Diaminobenzidine reagent was used to develop the immunoblots.
Estimation of glutathione
Total reduced glutathione (GSH) was determined by the method of Moron et al.[10]. Five percent TCA was added to HepG2 (1×106 cells). The precipitate was removed by centrifugation. To an aliquot of the supernatant, 2mL of DTNB reagent was added to make a final volume of 3 mL. The absorbance was read at 412 nm against a blank containing TCA instead of sample. Aliquots of the standard solution were treated similarly. The amount of GSH was expressed as nmoles/106 cells.
Statistical analysis
Values were expressed as mean ± SD. Difference between the drug-treated group and control group was analyzed with independent sample-‘t’ test using SPSS 7.5 student version. P < 0.05 was considered statistically significant.
RESULTS
Trypan blue exclusion method
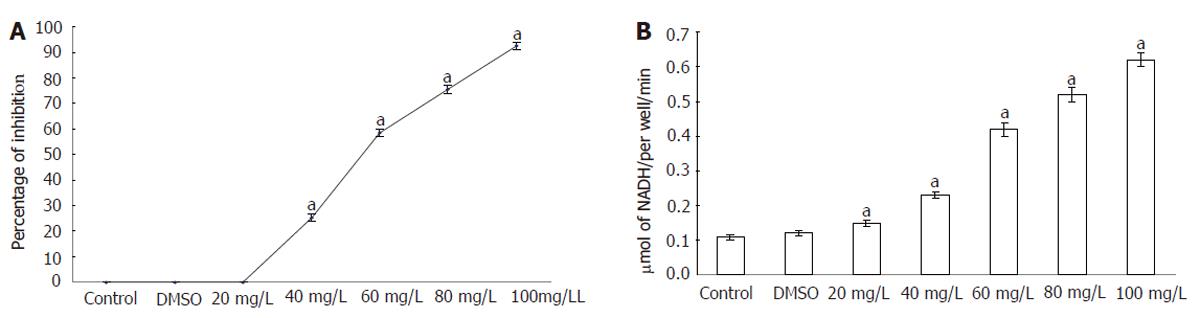
The cytotoxicity of T. arjuna to HepG2 cells was examined by trypan blue exclusion and LDH leakage assay. Figure 1 (A) shows the viability of control and T. arjuna-treated (20, 40, 60, 80 and100 mg/L) HepG2 cells. In the present study, the plant extract induced cytotoxicity to HepG2 cells in a concentration-dependent manner after 48 h of treatment. The results showed that treatment with T. arjuna could increase the number of apoptotic cells and decrease the number of normal HepG2 cells. Apoptotic cells were detached from the surface and suspended in the medium.
Figure 1 Viability (A) and LDH level (B) in control and HepG2 cells.
Cells were exposed to different concentrations of T. arjuna extract for 48 h and percentage of viable and dead cells was determined using trypan blue exclusion method. Results were expressed as the percent of total cell number and the percentage of LDH leakage was analyzed. Values are mean ± SD (n = 6). aP < 0.05 represents the statistical significance between control and T. arjuna treated HepG2 cells.
Lactate dehydrogenase leakage assay
The levels of LDH released into the medium of control and T. arjuna-treated (20, 40, 60, 80 and100 μg/mL) HepG2 cells are presented in Figure 1 (B). LDH activities were significantly elevated after 48 h of exposure to T. arjuna extract in the medium when compared to the control.
Light microscopic observation
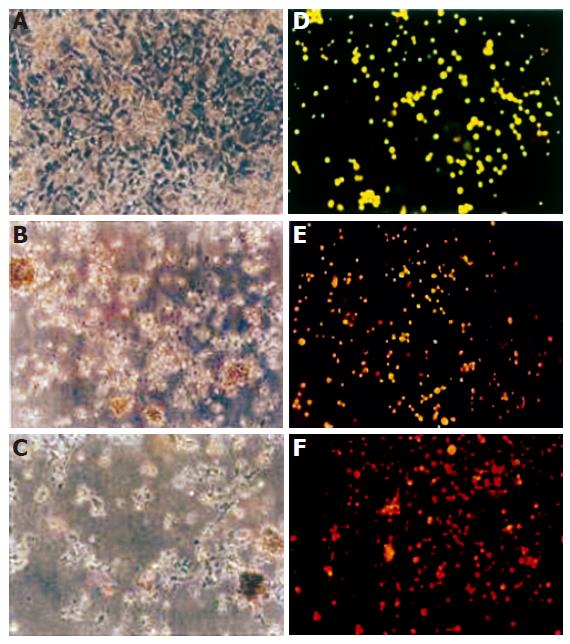
In T. arjuna-treated HepG2 cells, destruction of monolayer was observed, which was not seen in HepG2 cells not treated with it. T. arjuna treatment resulted in swelling and rounded morphology of HepG2 cells with condensed chromatin and their membrane also became crooked and vesicle shaped. Progressive structural alterations and reduction of HepG2 cell populations were observed [Figure 2 (A-C)].
Figure 2 Morphological changes of HepG2 cells under light and fluorescence microscope.
HepG2 cells were incubated for 48 h with T. arjuna extract. The medium was removed and the cells were rinsed and visualized under light microscope in control (A), 60 mg/LL (B), 100 mg/L (C) of T. arjuna extract. Cells were stained with ethidium bromide and acridine orange and observed under fluorescence microscope in control (D), 60 mg/L (E) and 100 mg/L (F) of T. arjuna extract.
Fluorescence microscopic observation
The morphological changes were observed in HepG2 cells treated with T. arjuna for 48 h. As shown in Figure 2 (D-F), normal live cells were bright green in color whereas T. arjuna-treated HepG2 cells were bright orange in color with condensed nuclei. Besides, normal nuclei showed chromatin with an organized structure, while apoptotic nuclei showed highly condensed chromatin in HepG2 cells treated with T. arjuna.
DNA fragmentation
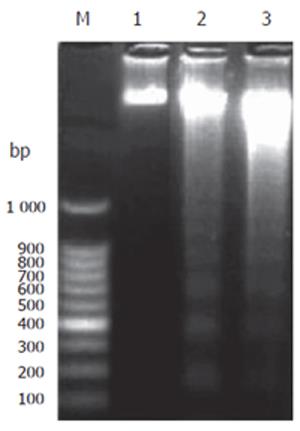
In the control, no fragmentation was observed in agarose gel. However, the signal appeared in T. arjuna extract (60 and 100 mg/L) treated HepG2 cells showed the fragmented laddering pattern of DNA, indicating the characteristics of apoptotic cells (Figure 3).
Figure 3 DNA fragmentation in HepG2 cells.
HepG2 cells were treated with T. arjuna extract and incubated for 48 h at the concentration of 60 and 100 mg/L, respectively. DNA fragmentation was carried out as described in MATERIALS AND METHODS. Lane 1: 100 bp DNA marker; Lane 2: control HepG2 cells without any treatment; Lanes 3 and 4: HepG2 cells treated with T. arjuna extract at the concentration of 60 and 100 μg/mL, respectively.
Expression of p53 and caspase-3 proteins
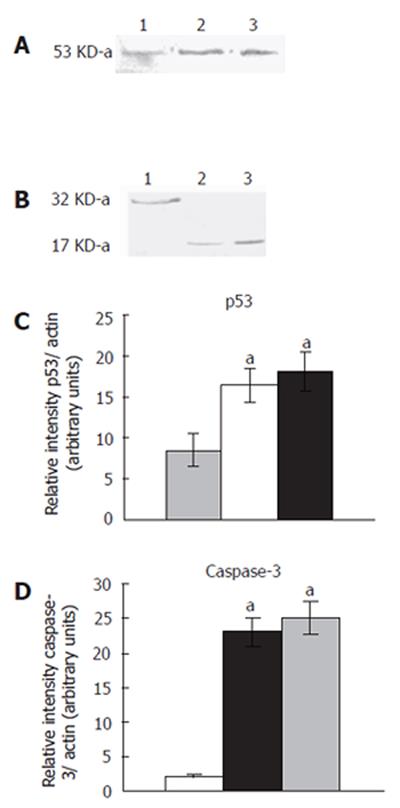
T. arjuna extract treatment increased the band intensity of 53-ku protein in HepG2 cells compared to the control (Figures 4A and 4B). The accumulation of p53 protein induced apoptosis in T. arjuna extract-treated HepG2 cells. On the contrary, HepG2 cells treated with T. arjuna extract (60 and 100 mg/L) reduced the intensity of 32-ku protein and appearance of 17-kDa protein measured by densitometric analysis (Figures 4C and 4D).This results indicated that T. arjuna treatment stimulated the proteolytic cleavage of caspase-3 protein and initiated the apoptosis pathway in HepG2 cells.
Figure 4 Expression of p53 and caspase-3 proteins.
Western blotting analysis of p53 and caspase-3 in HepG2 cells cultured for 48 h in the presence and absence of T. arjuna extract at the concentration 60 and 100 mg/L was carried out. Cell lysates were prepared as described in MATERIALS AND METHODS, and resolved by 12% SDS-PAGE, immunoblotted and detected using specific antibodies which recognized p53 (A) and caspase-3 (B). Densitometric analysis of the p53 (C) and caspase-3 band (D) was performed. Each value is mean ± SD of three observations. aP < 0.05 represents the statistical significance between control and T. arjuna extract-treated HepG2 cells.
GSH levels
The level of GSH content in control was 24.38 ± 1.37 n moles/106 cells. After treatment with T. arjuna (60 and 100 mg/L) the GSH level decreased to 18.57 ± 1.56 n moles/106 cells and 16.62 ± 1.46 n moles/106 cells, respectively. There was a significant difference between the T. arjuna extract-treated group and control group.
DISCUSSION
In the present investigation, T. arjuna extract markedly reduced the cell viability in a concentration-dependent manner. The suppression of cell proliferation induced by this extract may be due to the induction of cell death. Thus, the inhibitory activity of T. arjuna extract provides evidence for the in vitro cytotoxicity.
Recent studies suggest that LDH is a more reliable and accurate marker of cytotoxicity, because damaged cells are fragmented completely during the course of prolonged incubation with substances [7]. In the present study, the LDH leakage increased significantly in T. arjuna extract-treated HepG2 cells when compared to the control cells. Extensive reports have documented on medicinal plant extract-induced cytotoxicity to cancer cells. It was reported that the acetone and methanolic extracts of T. arjuna inhibit the growth of transformed cells such as osteosarcoma cells (U2OS) and glioblastoma cells (U251) in vitro at the concentration of 30 and 60 mg/L[11]. Thus, the inhibition of HepG2 cells strongly proves the cytotoxic nature of T. arjuna extract. Pasquini et al [12] also found that various fractions of T. arjuna inhibit the mutagenicity of 4-nitroquinoline-4-oxide in the salmonella/microsome test, suggesting that the inhibition may be due to the presence of antitumor promoters of the plant extract. Hence, the LDH leakage in HepG2 cells may be due to the cytotoxic nature of the plant extract and confirms its antitumor activity.
Light microscopic observation of T. arjuna extract-treated HepG2 cells at the concentrations of 60 and 100 mg/L after 48 h of exposure showed the typical morphological features of apoptosis in HepG2 cells. The morphological changes observed were reduction in cell volume, cell shrinkage, reduction in chromatin condensation and formation of cytoplasmic blebs. However, the control HepG2 cells had a higher confluency of monolayer.
Fluorescence microscopic study showed apoptosis in T. arjuna extract-treated HepG2 cells after exposed to the concentrations of 60 and 100 mg/L. In the present investigation, control HepG2 cells were stained green in color with acridine orange, because of cell membrane integrity. On the other hand, the drug-treated HepG2 cells were stained bright orange in color with ethidium bromide. This may be due to loss of cell membrane integrity and apoptotic nature of the plant extract.
Apoptotic cells often produce a unique ladder composed of nucleotide fragments at an interval of 180-200 base pairs, which can be visualized by DNA-agarose gel electrophoresis. It is generally assumed that the toxicity of antitumor drugs is the consequences of their ability to cause genomic DNA damage in cancer cells [13]. In this context, Bai and Cederbaum[14] reported that many chemotherapeutic drugs, including Vp16 and mitomicin C, induce cells to undergo apoptosis through damage of nuclear DNA. In the present study, DNA ladders appeared in T. arjuna extract-treated HepG2 cells after exposed to the concentrations of 60 and 100 μg/mL for 48 h. However, the control HepG2 cells did not show any DNA fragmentation. In general, cytotoxic drugs induce a massive breakage of DNA into oligonucleosome fragments. The degradation of DNA down to oligonucleosomal fragments is a late event of apoptosis [15]. Thus, the T. arjuna extract-induces DNA damage in HepG2 cells and thereby causes apoptosis.
It is well known that p53 acts as a guardian of the genome, and is one of the major factors controlling cell proliferation, growth and transformation. The p53 tumor suppressor gene is mutated in over 50% of human cancers, and the oncogenetic activity of p53 mutation is due to its ability to interfere with p53-dependent apoptosis by a dominant negative mechanism[16]. Tumor suppressor gene p53 is one of the critical genes regulating the onset of DNA replication around G1/S boundary. Also, p53-mediated tumor suppression appears to be critical for therapeutic potential in the treatment of tumors[17]. p53 is implicated in cell cycle control, DNA repair and apoptosis [18,19]. Thus, apoptotic modulators may be promising agents for treatment of HCC. p53 contributes to apoptosis induced by a variety of cellular stresses, including DNA damage, oxidative stress and chemotherapeutic drugs[20].
Activation of p53 is often deducted with natural chemotherapeutic agents and p53 negative tumors are generally less sensitive or even insensitive to these agents. In the present study, T. arjuna extract-treated HepG2 cells showed upstream regulation of p53 protein expression after exposed to the concentrations of 60 and 100 mg/L for 48 h. Hence, T. arjuna extract may possibly enhance the susceptibility of HepG2 cells to apoptosis by attenuating the tumor suppressor protein.
In cells undergoing apoptosis, there is a activation of proteases known as caspases, which have an obligatory cystein residue within the active site and cleave peptides adjacent to an aspartic acid residue[21]. Caspase cascade has been demonstrated to be involved in apoptotic signal transduction and execution[22]. Human caspases 1-10 have been described and activation of the caspase cascade is involved in chemical-induced apoptosis[23], including degradation of DNA repair enzyme poly ADP ribosepolymerase [24] and DNA-dependent protein kinase as well as cleavage of chromatin at inter-nucleosomal sites mediated by caspase-activated DNase[25]. Generally, caspases are present as inactive proenzymes, most of which are activated by proteolytic cleavage. Caspases-8, -9 and -3 are situated at pivotal junctions in apoptotic pathways[26]. Caspase-3 may then cleave vital cellular proteins or activate additional caspases by proteolytic cleavage. In the present investigation, T. arjuna extract-treated HepG2 cells showed a low intensity of 32-ku protein band and 17-ku protein band was observed. According to Busihardjo et al [27], caspase-3 usually exists as an inactive procaspase-3 that becomes proteolytically activated by multiple cleavages of its 32-ku precursor to generate the 20/11 or 17/11 ku active forms in cells undergoing apoptosis. Thus, the appearance of 17-ku protein may be one of the active forms of caspase-3 protein. It may be due to proteolytic cleavage of inactive caspase-3 induced by the plant extract. This occurs before the further activation of caspase-3-mediated apoptosis. Recent studies have also suggested that the proteolytic degradation of specific substrates is responsible for many of the morphological features of apoptosis[28,29].
It was reported that certain products from plants can induce apoptosis in neoplastic cells but not in normal cells [30,31]. Studies have shown that cytotoxic effect of the phenolic compounds on different tumors is mediated through apoptosis. For instance, gallic acid selectively induces cell death in various transformed cell lines such as PLC/PRF/5 (human hepatoma), HL-60, RG (human promyelocytic leukemia) and P-388D1 (mouse lymphoid neoplasma) the[32].
It has been reported that several putative compounds isolated from T. arjuna such as tannic acid, especially flavonoide (luteolin) are potent antitumor promoters and inhibitors of a series of solid tumors and leukemia [33,34]. Nagpal et al [11] have reported that the growth of osteosarcoma (U2OS) and glioblastoma (U251) is inhibited after treated with T. arjuna extract, suggesting that the induction of apoptosis in HepG2 cells may be due to the antitumor property of T. arjuna extract.
Numerous data indicate that intracellular oxidative metabolites play a significant role in the regulation of apoptosis. The reduced tripeptide GSH is a hydroxyl radical and a singlet oxygen scavenger, participating in a wide range of cellular functions such as protein and DNA synthesis, intermediary metabolism and transport [35]. The maintenance of GSH levels and the reducing environment of cells are crucial [36]. Depletion of GSH leads to increased accumulation of lipid peroxides and loss of cell viability [37]. It is known that the toxicity of antitumor drugs may largely depend on the intracellular level of reduced GSH. Because GSH is the main antioxidant system in cells, a possible explanation is that GSH depletion facilitates reactive oxygen species (ROS) accumulation in cells treated with antitumor drugs [38], which in turn increases their lethality. In the present study, the level of GSH significantly decreased in T. arjuna extract-treated HepG2 cells at the concentration of 60 and 100 μg/mL, indicating that the decrease in GSH levels may initiate redox imbalance in HepG2 cells and subsequently induces apoptosis. Depletion of GSH has been described for several agents such as oxidative and alkylating agents in various cell types [39]. Cells with reduced GSH levels either undergo apoptosis or become more sensitive to various death inducing agents[40,41] Liu et al.[42] also found that Salvia miliorrhiza inhibits human hepatoma HepG2 cell growth and induces apoptosis involving intracellular GSH depletion. Hall [43] has demonstrated that onset of apoptosis is associated with a fall of intracellular GSH in different cellular systems. Loss of GSH is tightly coupled with a number of downstream events in apoptosis [44].
Studies in a variety of cell types suggest that cancer chemotherapeutic drugs induce tumor cell apoptosis in part by increasing the formation of ROS. According to Simizu et al.[45], some anticancer agents, including vinblastin and camptothecin, induce cell apoptosis with the generation of ROS. Previous reports suggest that quercetin and related phenolic compounds containing a catechol moiety in their chemical structures are oxidized under certain in vitro conditions, resulting in lipid peroxidation [46,47]. Accordingly, it is speculated that T. arjuna extract may induce ROS production in HepG2 cells and subsequently causes apoptosis. However, ROS may not necessarily be the direct factor to cause apoptosis induced by the drug, but intracellular ROS may modulate the genes involved in apoptosis, which may regulate apoptosis. Thus, T. arjuna extract-induced oxidative stress is upstream of signaling events that might alter the pro and antiapoptotic balance in HepG2 cells. Phenolic compounds are generally known to show not only their antioxidant effects but also pro-oxidant actions under in vitro conditions. Hence, it is possible that treatment of HepG2 cells with T. arjuna extract can deplete the GSH levels and promote oxidation induction, which switches the mode of death via apoptosis. Therefore, the cytotoxic action of this drug may be attributed to its pro-oxidant action on the cells. This may be able to account for the discrepancy between in vitro cytotoxicity and in vivo antitumor activities of T. arjuna extract.
In conclusion, T. arjuna extract-has profound effects on human hepatoma cellline HepG2 and exhibits its cytotoxicity to these cells and the cell death is mediated by apoptosis. The mechanism of apoptosis may be accumulation of p53 protein and proteolytic cleavage of caspase-3 protein. In addition to these, GSH depletion may also play a role in apoptosis through redox imbalance in HepG2 cells. Thus, our results provide the basis for further in vivo and clinical research.