Published online Feb 7, 2006. doi: 10.3748/wjg.v12.i5.809
Revised: May 19, 2005
Accepted: July 15, 2005
Published online: February 7, 2006
Hereditary non-polyposis colorectal carcinoma (HNPCC) is an autosomal dominant disorder associated with colorectal and endometrial cancer and a range of other tumor types. Germline mutations in the DNA mismatch repair (MMR) genes, particularly MLH1, MSH2, and MSH6, underlie this disorder. The vast majority of these HNPCC-associated mutations have been proven, or assumed, given the family history of cancer, to be transmitted through several generations. To the best of our knowledge, only a single case of a de novo germline MMR gene mutation (in MSH2) has been reported till now. Here, we report a patient with a de novo mutation in MLH1. We identified a MLH1 Q701X truncating mutation in the blood lymphocytes of a male who had been diagnosed with rectal cancer at the age of 35. His family history of cancer was negative for the first- and second-degree relatives. The mutation could not be detected in the patient’s parents and sibling and paternity was confirmed with a set of highly polymorphic markers. Non-penetrance and small family size is the common explanation of verified negative family histories of cancer in patients with a germline MMR gene mutation. However, in addition to some cases explained by non-paternity, de novo germline mutations should be considered as a possible explanation as well. As guidelines that stress not to restrict MMR gene mutation testing to patients with a positive family history are more widely introduced, more cases of de novo MMR gene germline mutations may be revealed.
-
Citation: Stulp RP, Vos YJ, Mol B, Karrenbeld A, Raad M, Mijle HJVD, Sijmons RH. First report of a
de novo germline mutation in theMLH1 gene. World J Gastroenterol 2006; 12(5): 809-811 - URL: https://www.wjgnet.com/1007-9327/full/v12/i5/809.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i5.809
Hereditary non-polyposis colorectal carcinoma (HNPCC) is a genetic disorder caused by germline mutations in DNA mismatch repair (MMR) genes, especially MLH1, MSH2 and MSH6. HNPCC is characterized clinically by the early-onset colorectal and endometrial cancer as well as a range of other tumor types. In contrast to familial adenomatous polyposis (FAP), HNPCC lacks an easily recognizable clinical phenotype in individual patients. The original and revised Amsterdam criteria (AC and AC II) introduced to clinically define HNPCC therefore include specific characteristics of the family history that should be met[1,2]. However, it has become clear that HNPCC patients may have a negative family history. Small family sizes and less than 100% penetrance of the germline mutations involved are its most common explanation. Here, we report the rare phenomenon of a de novo germline MMR gene mutation as the cause of a negative family history in a patient with hereditary non-polyposis colorectal carcinoma.
A 35-year-old male had been referred to our department for genetic testing and counseling because he had very recently been diagnosed with a T4N0M0 rectal adenocarcinoma. Family history of cancer was negative for the first- and second-degree relatives of the patient. The patient’s mother, father, and sibling (aged 60, 61, and 38 years, respectively) were healthy.
As part of an ongoing study on the genetics of colorectal cancer, testing for tumor microsatellite instability (MSI), tumor immunohistochemistry for the MLH1, MSH2 and MSH6 proteins as well as mutation analysis of MLH1, MSH2 and MSH6 are offered to all the patients with colorectal cancer diagnosed before the age of 50 who are referred to our department for genetic testing and counseling.
DNA was extracted from formalin-fixed and paraffin-embedded tumor and non-tumor tissues as described previously[3]. Subsequent MSI analysis was performed using criteria and primers as described in the Bethesda guidelines[4]. PCR products were analyzed on 60 g/L denaturing polyacrylamide gels with an LKB ALF DNA sequencer. For data analysis, the DNA fragment analyzer (Pharmacia) was used.
Immunohistochemistry for MLH1, MSH2, and MSH6 protein expression was carried out as described previously[5]. Protein expression in normal tissue surrounding the cancer served as internal control.
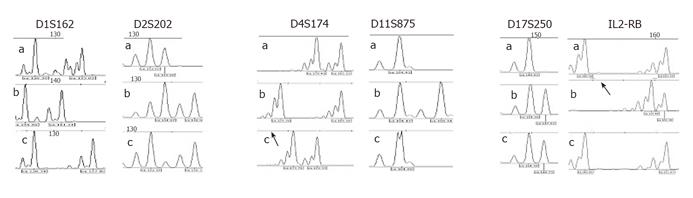
Mutation analysis of the MLH1 and MSH2 genes in DNA obtained from peripheral blood lymphocytes was carried out using denaturing gradient gel electrophoresis (DGGE), followed in case of aberrant patterns by direct sequencing of independently amplified PCR products as previously described[6,7]. To investigate paternity, a panel of six highly polymorphic markers was analyzed with PCR using markers D1S162, D2S202, D4S174, D11S875, D17S250, and IL2-RB in the patient and his parents. PCR products were analyzed on 60 g/L denaturing polyacrylamide gels with an LKB ALF DNA sequencer. For data analysis, the DNA fragment analyzer (Pharmacia) was used.
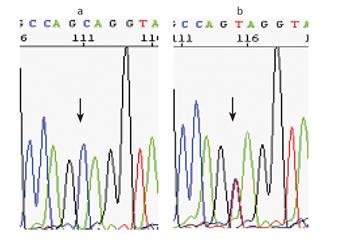
The tumor of the patient revealed microsatellite instability (MSI-H), as well as loss of immunohistochemical staining for the MLH1 protein with a positive internal control. Mutation analysis revealed a C to T substitution at nucleotide 2 101 in exon 18 of the MLH1 gene, leading to an early stop codon (Gln701Stop). Neither the father nor the mother carried the mutation identified in their son (Figures 1 and 2). All six tested markers were in concordance with paternity (likelihood: >99.9%) (Figure 3)
Although the Q701X MLH1 mutation has not been reported before in either the Human Gene Mutation Database (HGMD) and the ICG-HNPCC database, we have no reason to doubt its pathogenicity given its truncating nature and the findings of MSI-H and loss of staining for the MLH1 protein in the patient’s tumor[8,9].
De novo germline mutations of the MMR genes in HNPCC are rare. To the best of our knowledge, only one single case, involving MSH2, has been reported so far[10]. We have identified a de novo germline mutation in the MLH1 gene. The classification “germline” mutation is generally used in oncogenetics to distinguish mutations that can be transmitted to the offspring from the somatic mutations occurring locally in tissues and which may lead to local tumor development. For practical purposes, we therefore used the “germline” label. Strictly speaking, mutations classified as de novo germline mutations should refer to a single mutation occurring in a single germcell. Clinically, these often cannot be distinguished from an early postzygotic mutational event in the patient, or from germline mosaicism or somatic mosaicism extending to extra-gonadal tissues in the parents. We did not have the opportunity to test multiple tissues in our patient or his parents for the mutation. The patient’s single sibling was offered mutation analysis and was shown to carry the wild-type MLH1 alleles. Therefore, we lack proof of the germline nature of the de novo mutation reported by us and the same is true for the MSH2 case published earlier by Kraus et al [10]. In clinical practice, this lack of proof does not lead to any obvious problems. One will simply discuss with the patient the fact that his or her siblings have a, probably small, risk to carry the mutation as well and one will offer to test them for it.
Non-penetrance and small family size is the common explanation of verified negative family histories of cancer in patients with a germline MMR gene mutation. However, in addition to some cases due to non-paternity, de novo“germline” mutations should be considered as a possible explanation as well. The percentage of these mutations in HNPCC remains to be established. As guidelines (e.g., the Bethesda guidelines)[4] that stress not to limit MMR gene mutation testing to patients with a positive family history are more widely introduced, more cases of de novo“germline” MMR mutations are yet to be revealed.
S- Editor Kumar M and Guo SY L- Editor Elsevier HK E- Editor Cao L
| 1. | Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on HNPCC. Anticancer Res. 1994;14:1661-1664. [PubMed] |
| 2. | Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1688] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 3. | Goelz SE, Hamilton SR, Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun. 1985;130:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 553] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 4. | Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2154] [Cited by in RCA: 2218] [Article Influence: 105.6] [Reference Citation Analysis (1)] |
| 5. | Berends MJ, Wu Y, Sijmons RH, Mensink RG, van der Sluis T, Hordijk-Hos JM, de Vries EG, Hollema H, Karrenbeld A, Buys CH. Molecular and clinical characteristics of MSH6 variants: an analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 210] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 6. | Wu Y, Nyström-Lahti M, Osinga J, Looman MW, Peltomäki P, Aaltonen LA, de la Chapelle A, Hofstra RM, Buys CH. MSH2 and MLH1 mutations in sporadic replication error-positive colorectal carcinoma as assessed by two-dimensional DNA electrophoresis. Genes Chromosomes Cancer. 1997;18:269-278. [PubMed] |
| 7. | Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, van Der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65:1291-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 222] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 8. | Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1346] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 9. | Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269-276. [PubMed] |
| 10. | Kraus C, Kastl S, Günther K, Klessinger S, Hohenberger W, Ballhausen WG. A proven de novo germline mutation in HNPCC. J Med Genet. 1999;36:919-921. [PubMed] |