Published online Dec 28, 2006. doi: 10.3748/wjg.v12.i48.7758
Revised: September 28, 2006
Accepted: November 29, 2006
Published online: December 28, 2006
AIM: To investigate the glutathione-S-transferase M1 (GSTM1) polymorphisms in three Chinese minorities, Kazakh, Uygur, and Tajik; and the pathological significance of GSTM1 polymorphisms in esophageal carcinogenesis in Kazakh.
METHODS: A total of 1121 blood samples (442 males and 679 females) were obtained from healthy Kazakh (654), Uygur (412) and Tajik (55). Primary esophageal squamous cell cancer (ESCC) tissues from Kazakh were obtained from 116 patients who underwent surgery. GSTM1 polymorphisms were analyzed by a combined approach of PCR and electrophoresis techniques.
RESULTS: GSTM1 null genotype was found in 62.63% Uygur, 50.91% Tajik and 47.40% Kazakh. A significantly higher frequency of GSTM1 null genotype in Uygur was observed compared with Kazakh (OR: 1.859, 95% CI: 1.445 -2.391, χ2 = 23.71, P = 0.000). In addition, GSTM1 null genotype was found in 23.53% of well-differentiated ESCC in Kazakh, in 49.23% of poorly differentiated ESCC, with a significant difference (OR: 3.152, 95% CI: 1.403-7.080, χ2 = 8.018, P = 0.007).
CONCLUSION: There is a marked difference in the frequency of common GSTM1 null genotype between Uygur and Kazakh. GSTM1 null genotype is associated with differentiation of ESCC in Kazakh.
- Citation: Lu XM, Yang T, Xu SY, Wen H, Wang X, Ren ZH, Zhang Y, Wang W. Glutathione-S-transferase M1 polymorphisms on the susceptibility to esophageal cancer among three Chinese minorities: Kazakh, Tajik and Uygur. World J Gastroenterol 2006; 12(48): 7758-7761
- URL: https://www.wjgnet.com/1007-9327/full/v12/i48/7758.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i48.7758
Esophageal cancer (EC) is the sixth leading cause of cancer mortality worldwide[1]. The incidence of EC is highly variable in different populations, with more than a 50-fold difference between the high- and low-risk ethic groups[2,3]. For example, Turkomans in northeastern Iran are considered to be a very high-risk group, with age standardized prevalence (ASR) of over 100/100 000 for both men and women; whereas the prevalence of EC in pure Zoroastrian Persians in Iran and India are known to be considerably low, with ASRs of 3-7/100 000.
Epidemiological studies have identified several high EC incidence areas, such as the western and northern parts of China[4], certain areas of France and Brazil[5]. In Xinjiang Uygur Autonomous Region of China, there are thirteen minority ethnical groups (Uygur, Han, Hazakh, Tajik, Hui, Uzbek, Kerkez, Man, Mongolia, Tatar, Darur, Xibo, and Russian), who have lived there since ancient times. Uygur, Hazakh and Tajik are the major residents among those minorities with populations of 8 million, 2 million, and 40 thousand, respectively. Although they are all Muslims and have certain similarities in their life styles, the morbidities of EC among them are quite different. The incidence of EC in Kazakh is highest among all ethnics in Xinjiang, with an age-adjusted mortality of 90.7/100 000, significantly higher than that in Uygur (23.4/100 000) and almost 18-fold higher than that in Tajik (5.13/100 000)[4].
Glutathione S-transferases (GSTs) constitute a superfamily of ubiquitous multifunctional enzymes, which play a key role in cellular detoxification and protection of macromolecules from being attacked by reactive electrophiles[6]. GSTs catalyze the conjugation of tripeptide glutathione (GSH) to a wide variety of exogenous and endogenous chemicals with electrophilic functional groups (e.g. products of oxidative stress, environmental pollutants, and carcinogens), thereby neutralizing their electrophilic sites, and rendering the products more water-soluble[7]. Based on sequence homology and immunological cross-reactivity, human cytosolic GSTs have been grouped into seven families, designated GST Alpha, Mu, Pi, Sigma, Omega, Theta, and Zeta[8]. The GSTs presumably arise from a single common ancestor and their substrate specificity and diversity have been reshaped by gene duplication, recombination and mutation.
There are marked intra- and inter-ethnic differences in the frequencies of common GST mutations[9,10]. For example, the distribution of GSTM1 genotype frequencies in Indian is significantly different from that in Chinese[11]. GSTM1 polymorphisms have been considered as a risk factor for EC development in a number of studies; however the overall results of such studies are inconsistent[12,13]. Up to date, data on genetic analysis of GSTM1 in Uygur, Tajik are lacking[14], and the correlation between GSTM1 polymorphisms and high incidence of EC in Kazakh has not been clarified.
The present study aimed to investigate the GSTM1 polymorphisms in healthy Kazakh, Uygur, and Tajik; and to explore the pathological significance of GSTM1 polymorphisms in esophageal carcinogenesis.
A total of 1121 blood samples was collected from healthy Kazakh (269 males and 385 females; age 35-65 years, mean 46.5 years), Uygur (146 males and 266 females; age 30-68 years, mean 45.5 years) and Tajik (27 males and 28 females; age 32-70 years, mean 47.5 years). All subjects from north-western of Xinjiang received clinical and biochemical assessments before entering this study and none of them has a clinical or family history of EC. Specimens of 116 primary EC tissues from Kazakh (84 males and 32 females; age 42-76 years, mean 55.5 years), with histological confirmation of primary ESCC, was recruited from two hospitals in Xinjiang from July 1999 to June 2004.
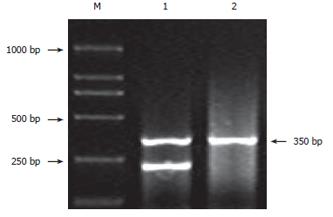
DNAs from healthy controls were extracted from peripheral leukocytes using the classical phenol-chloroform extraction method[15]. Genomic DNA in cancer tissue embedded by paraffin was prepared by proteinase K digestion and phenol/chloroform extraction, followed by ethanol precipitation, as described by Diffenbach[16]. The purity and concentration of DNA was examined by ultraviolet densitometry. GSTM1 genotyping for gene deletion was performed by PCR using primers 5’-GAACTCCCTGAAAAGCTAAAGC-3’ and 5’-GTTGGGCTCAAATATACGGTGG-3’[17], which produced a 219 bp product. At the same time, β-globin gene was amplified, resulting in a 350 bp product as an internal control. PCR was performed in a reaction mixture of 20 μL containing 100 ng sample DNA, 10 mmol/L Tris-HCl, 50 mmol/L KCl, 1.5 mmol/L MgCl2 pH 8.4, 0.1 mmol/L of each dNTP and 1.25 U Taq polymerase. After initial denaturation for 5 min at 94°C, 35 cycles were performed at 94°C for 30 s (denaturation), at 63°C for 30 s (annealing) and at 72°C for 30 s (extension), followed by a final step for 5 min at 72°C. The amplified products were visualized by electrophoresis in ethidium-bromide-stained 1.5% agarose gel in TBE buffer. For genotype of GSTM1 deletion, no amplified product was observed except the band of β-globin gene.
Chi-square test was used to examine the correlation between the GSTM1 polymorphism among three healthy ethnics, and association of GSTM1 polymorphisms with differentiation of ESCC in Kazakh with SPSS software (11.0). Odds ratios (ORs) and 95% confidence intervals (CIs) of different variables among groups were calculated.
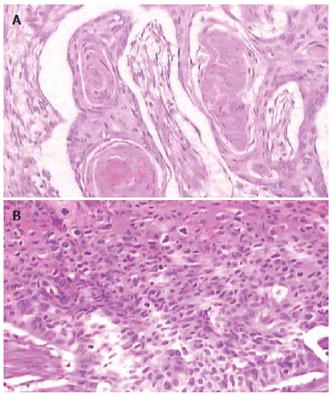
Histological confirmation of primary ESCC including well-differentiated and poorly-differentiated are shown in Figure 1.
Figure 2 shows the PCR-amplified fragment of GSTM1. Genotype data for GSTM1 in the three ethnics are summarized in Table 1. The frequency of GSTM1 null mutation in Kazakh was significantly lower than that in Uygur (OR:1.859, 95% CI: 1.445-2.391, χ2 = 23.71, P = 0.000, P < 0.05). There was no significant difference in the frequency of the GSTM1 null genotype between Uygur (62.63%) and Tajik (50.91%) (χ2 = 2.804, P > 0.05), and there was no significant difference between the Kazakh (47.40%) and Tajikic either (χ2 = 0.250, P > 0.05). In addition, no significant difference of GSTM1 null polymorphisms between the two genders of each ethnical group was observed.
| Ethnic | GSTM1 | OR (95% CI) | P | |
| Null [n (%)] | Present [n (%)] | |||
| Kazakh | ||||
| Male | 130 (48.33) | 139 (51.67) | ||
| Female | 180 (46.75) | 205 (53.25) | ||
| Total | 310 (47.40) | 344 (52.60) | ||
| 11.859 (1.445-2.391) | 0.000 | |||
| Uygur | ||||
| Male | 99 (67.81) | 47 (32.19) | ||
| Female | 159 (59.77) | 107 (40.23) | ||
| Total | 258 (62.63) | 154 (37.37) | ||
| 20.619 (0.352-1.809) | 0.106 | |||
| Tajik | ||||
| Male | 13 (50.00) | 13 (50.00) | ||
| Female | 15 (51.72) | 14 (48.28) | ||
| Total | 28 (50.91) | 27 (49.09) | ||
| 31.151 (0.664-1.996) | 0.674 | |||
There was a significant difference in the frequency of the GSTM1 null genotype between well-differentiation (high grade) (76.47%) and poor-differentiation (low grade) group (50.77%) of EC of Kazakh (OR 3.152, 95% CI 1.403-7.080, χ2 = 8.018, P < 0.05) (Table 2).
| ESCC grade | GSTM1 | OR (95% CI) | P | |
| Null [n (%)] | Present [n (%)] | |||
| High | 12 (23.53) | 39 (76.47 ) | ||
| Low | 32 (49.23) | 33 (50.77) | 3.152 (1.403-7.080) | 0.007 |
The odds ratio of GSTM1 null genotype of Kazakh people with lowly differentiated ESCC was 3.152-fold higher than those people with highly differentiated ESCC.
In this study, we investigated differences in the prevalence of GSTM1 null genotypes in three ethnical groups, Kazakh, Tajik and Uygur, in Xinjiang. As far as we know, we are the first to report the frequency of GSTM1 null genotype in Tajik. GSTM1 null genotype in Uygur in Xinjiang has a similar frequency when compared with Zoroastrians Iranian[18] and Han Chinese[19,20].
The study showed the association of GSTM1 null genotype with ESCC differentiation in Kazakh, suggesting the involvement of GSTM1 null genotype in the development of ESCC. Differences in the risk of EC development between high- and low-risk populations may partly be attributed to the genetic make-up of the populations, reflected by their different susceptibility to EC. GSTM1 encoding metabolic enzymes, the alteration in expression and function of which may increase or decrease carcinogen activation/detoxication, expressed as different phenotypes with different cancer risk[21-23]. Homozygous deletions of such genes, called GSTM1 null genotypes, result in the phenotype of no enzyme activity[24]. Individuals with null genotypes of GSTM1 are reported at high risk for developing several types of cancers, e.g. breast, lung, cervix[25-27] and bladder cancers[28-32]. However, the frequency of GSTM1 null genotype was low in Kazakh with high risk to EC, suggesting that the lack of the null allele or the other genes may play roles in carcinogenesis of ESCC by different mechanisms or via different pathways, from that of the reported breast, lung, cervix and bladder cancers. This large sample study on 654 of healthy Kazakh individuals and our previous genotyping results[14] have confirmed this contradictory finding of low frequency GSTM1 null genotype among Kazakh with a high susceptibility to ESCC.
In conclusion, there are different frequencies of GSTM1 null genotype among Uygur, Tajik and Kazakh, however, a significant difference is only observed between Uygur and Kazakh. The GSTM1 null genotype may play a role in the carcinogenesis and progress of ESCC.
We thank those people who provided the blood samples of Kazakh, Tajik and Uygur individuals.
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2651] [Cited by in RCA: 2599] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 2. | Munoz N, Day NE. Esophageal cancer. Cancer Epidemiology and Prevention. 2nd ed. New York: Oxford University Press 1996; 681-706. |
| 3. | Sadjadi A, Malekzadeh R, Derakhshan MH, Sepehr A, Nouraie M, Sotoudeh M, Yazdanbod A, Shokoohi B, Mashayekhi A, Arshi S. Cancer occurrence in Ardabil: results of a population-based cancer registry from Iran. Int J Cancer. 2003;107:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 140] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 4. | Zhang YM. The distribution of esophageal cancer in Xinjiang. Xinjiang Yixueyuan Xuebao. 1988;11:139-144. |
| 5. | Pickens A, Orringer MB. Geographical distribution and racial disparity in esophageal cancer. Ann Thorac Surg. 2003;76:S1367-S1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 125] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 6. | Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 586] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 7. | Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2409] [Cited by in RCA: 2419] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 8. | Board PG, Coggan M, Chelvanayagam G, Easteal S, Jermiin LS, Schulte GK, Danley DE, Hoth LR, Griffor MC, Kamath AV. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J Biol Chem. 2000;275:24798-24806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 539] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 9. | Mondal BC, Paria N, Majumdar S, Chandra S, Mukhopadhyay A, Chaudhuri U, Dasgupta UB. Glutathione S-transferase M1 and T1 null genotype frequency in chronic myeloid leukaemia. Eur J Cancer Prev. 2005;14:281-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Ntais C, Polycarpou A, Ioannidis JP. Association of GSTM1, GSTT1, and GSTP1 gene polymorphisms with the risk of prostate cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2005;14:176-181. [PubMed] |
| 11. | Chowbay B, Zhou S, Lee EJ. An interethnic comparison of polymorphisms of the genes encoding drug-metabolizing enzymes and drug transporters: experience in Singapore. Drug Metab Rev. 2005;37:327-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Yang CX, Matsuo K, Wang ZM, Tajima K. Phase I/II enzyme gene polymorphisms and esophageal cancer risk: a meta-analysis of the literature. World J Gastroenterol. 2005;11:2531-2538. [PubMed] |
| 13. | Wang AH, Sun CS, Li LS, Huang JY, Chen QS, Xu DZ. Genetic susceptibility and environmental factors of esophageal cancer in Xi'an. World J Gastroenterol. 2004;10:940-944. [PubMed] |
| 14. | Lu XM, Zhang YM, Lin RY, Arzi G, Wang X, Zhang YL, Zhang Y, Wang Y, Wen H. Relationship between genetic polymorphisms of metabolizing enzymes CYP2E1, GSTM1 and Kazakh's esophageal squamous cell cancer in Xinjiang, China. World J Gastroenterol. 2005;11:3651-3654. [PubMed] |
| 15. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40518] [Cited by in RCA: 39099] [Article Influence: 1028.9] [Reference Citation Analysis (0)] |
| 16. | Greer CE, Whee Le CM, Manos MM. PCR Primer A Laboratory Manual. Cold Spring Harbor Laboratory Press. New York: Oxford University Press 1995; 64-69. |
| 17. | Arand M, Mühlbauer R, Hengstler J, Jäger E, Fuchs J, Winkler L, Oesch F. A multiplex polymerase chain reaction protocol for the simultaneous analysis of the glutathione S-transferase GSTM1 and GSTT1 polymorphisms. Anal Biochem. 1996;236:184-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 231] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 18. | Sepehr A, Kamangar F, Abnet CC, Fahimi S, Pourshams A, Poustchi H, Zeinali S, Sotoudeh M, Islami F, Nasrollahzadeh D. Genetic polymorphisms in three Iranian populations with different risks of esophageal cancer, an ecologic comparison. Cancer Lett. 2004;213:195-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Tan W, Song N, Wang GQ, Liu Q, Tang HJ, Kadlubar FF, Lin DX. Impact of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1 on susceptibility to esophageal cancer among high-risk individuals in China. Cancer Epidemiol Biomarkers Prev. 2000;9:551-556. [PubMed] |
| 20. | Zhong SL, Zhou S, Chen X, Huang M. Rapid determination of common mutations in glutathione S-transferase gene by PCR-based methods in healthy Chinese. Clin Chim Acta. 2006;364:205-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Olshan AF, Weissler MC, Watson MA, Bell DA. GSTM1, GSTT1, GSTP1, CYP1A1, and NAT1 polymorphisms, tobacco use, and the risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:185-191. [PubMed] |
| 22. | Gajecka M, Rydzanicz M, Jaskula-Sztul R, Kujawski M, Szyfter W, Szyfter K. CYP1A1, CYP2D6, CYP2E1, NAT2, GSTM1 and GSTT1 polymorphisms or their combinations are associated with the increased risk of the laryngeal squamous cell carcinoma. Mutat Res. 2005;574:112-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Ueda M, Hung YC, Terai Y, Saito J, Nunobiki O, Noda S, Ueki M. Glutathione-S-transferase and p53 polymorphisms in cervical carcinogenesis. Gynecol Oncol. 2005;96:736-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300:271-276. [PubMed] |
| 25. | Chacko P, Joseph T, Mathew BS, Rajan B, Pillai MR. Role of xenobiotic metabolizing gene polymorphisms in breast cancer susceptibility and treatment outcome. Mutat Res. 2005;581:153-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | Pinarbasi H, Silig Y, Cetinkaya O, Seyfikli Z, Pinarbasi E. Strong association between the GSTM1-null genotype and lung cancer in a Turkish population. Cancer Genet Cytogenet. 2003;146:125-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | García-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, Tardón A, Serra C, Carrato A, García-Closas R. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 451] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 28. | Engel LS, Taioli E, Pfeiffer R, Garcia-Closas M, Marcus PM, Lan Q, Boffetta P, Vineis P, Autrup H, Bell DA. Pooled analysis and meta-analysis of glutathione S-transferase M1 and bladder cancer: a HuGE review. Am J Epidemiol. 2002;156:95-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 153] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 29. | Jong Jeong H, Jin Kim H, Young Seo I, Ju Kim H, Oh GJ, Cheon Chae S, Sik Lim J, Taeg Chung H, Joong Kim J. Association between glutathione S-transferase M1 and T1 polymorphisms and increased risk for bladder cancer in Korean smokers. Cancer Lett. 2003;202:193-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 30. | Saadat M, Farvardin-Jahromi M, Saadat H. Null genotype of glutathione S-transferase M1 is associated with senile cataract susceptibility in non-smoker females. Biochem Biophys Res Commun. 2004;319:1287-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Lee SA, Kim JW, Roh JW, Choi JY, Lee KM, Yoo KY, Song YS, Kang D. Genetic polymorphisms of GSTM1, p21, p53 and HPV infection with cervical cancer in Korean women. Gynecol Oncol. 2004;93:14-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | Ye Z, Song H. Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and meta-analysis. Eur J Cancer. 2005;41:980-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 112] [Article Influence: 5.6] [Reference Citation Analysis (0)] |