Published online Dec 21, 2006. doi: 10.3748/wjg.v12.i47.7626
Revised: October 1, 2006
Accepted: October 10, 2006
Published online: December 21, 2006
AIM: To study the action of hepatitis virus infection-associated genes at transcription level during liver regeneration (LR).
METHODS: Hepatitis virus infection-associated genes were obtained by collecting the data from databases and retrieving the correlated articles, and their expression changes in the regenerating rat liver were detected with the rat genome 230 2.0 array.
RESULTS: Eighty-eight genes were found to be associated with liver regeneration. The number of genes initially and totally expressed during initial LR [0.5-4 h after partial hepatectomy (PH)], transition from G0 to G1 (4-6 h after PH), cell proliferation (6-66 h after PH), cell differentiation and reorganization of structure-function (66-168 h after PH) was 37, 8, 48, 3 and 37, 26, 80, 57, respectively, indicating that the genes were mainly triggered at the early stage of LR (0.5-4 h after PH), and worked at different phases. These genes were classified into 5 types according to their expression similarity, namely 37 up-regulated, 9 predominantly up-regulated, 34 down-regulated, 6 predominantly down-regulated and 2 up/down-regulated genes. Their total up- and down-regulation frequencies were 359 and 149 during LR, indicating that the expression of most genes was enhanced, while the expression of a small number of genes was attenuated during LR. According to time relevance, they were classified into 12 groups (0.5 and 1 h, 2 and 4 h, 6 h, 8 and 12 h, 16 and 96 h, 18 and 24 h, 30 and 42 h, 36 and 48 h, 54 and 60 h, 66 and 72 h, 120 and 144 h, 168 h), demonstrating that the cellular physiological and biochemical activities during LR were fluctuated. According to expression changes of the genes, their expression patterns were classified into 23 types, suggesting that the cellular physiological and biochemical activities during LR were diverse and complicated.
CONCLUSION: The anti-virus infection capacity of regenerating liver can be enhanced and 88 genes play an important role in LR.
- Citation: Su LJ, Ding GW, Yang ZL, Zhang SB, Yang YX, Xu CS. Expression patterns and action analysis of genes associated with hepatitis virus infection during rat liver regeneration. World J Gastroenterol 2006; 12(47): 7626-7634
- URL: https://www.wjgnet.com/1007-9327/full/v12/i47/7626.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i47.7626
The liver can regenerate and precisely regulate its size. Hepatocytes maintain the ability to proliferate in response to hepatectomy, liver damage caused by viruses or chemicals, liver cell death, etc[1,2]. The remaining liver may compensate for the lost hepatic tissue in which the growth situation of regenerating liver can be accurately detected, and liver regeneration (LR) may stop at a proper time point[3,4]. The regeneration process is usually categorized based on hepatic physiological biochemical activities into four stages: initiation [0.5-4 h after partial hepatectomy (PH)], transition from G0 to G1 (0.5-4 h after PH), cell proliferation (6-66 h after PH), cell redifferentiation and reorganization of the architecture-function (66-168 h after PH)[5]. In this process, the cellular physiological and biochemical activities and the gene expression situations have a very sweeping change.
Hepatitis viruses causing hepatic injury and liver diseases[6] include hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV) and hepatitis E virus (HEV)[7]. HAV and HEV do not result in chronic hepatic inflammation because of their self-limited infection, whereas HBV, HCV and HDV cause both acute and chronic hepatic inflammation, and finally result in chronic hepatitis, cirrhosis or hepatoma[8-10]. In infection, viruses first bind specifically to receptors, then complete their replication cycle by many processes such as adsorbing, penetrating, shelling, biosynthesis, assembling and release. Viruses interact with cells, and alter the gene expression of host cells. The process during which the viruses are cleaned by the host immune system results in changes in physiology and pathology of the host[11,12]. It has been reported that liver tissues with hepatitis may appear inflammation, necrosis or hyperplasia, with which over 200 genes are associated[9,13].
Since hepatitis virus infection-associated genes have a wide variety of kinds and complicated functions, it is almost impossible to give insights into their action during liver regeneration at transcriptional level unless high-throughput gene expression profile analysis is performed[14-16]. Therefore, we used the rat genome 230 2.0 array containing 193 genes associated with hepatitis virus infection to detect the gene expression changes in the regenerating liver after 2/3 hepatectomy. A total of 88 genes were found to be associated with LR, and their expression changes, patterns and actions during liver regeneration were primarily analyzed.
Healthy SD rats weighing 200-250 g were obtained from the Animal Center of Henan Normal University. The rats were divided into groups at random, 6 rats in each group (Male: Female = 1:1). Partial hepatectomy (PH) was performed as previously described[17], the left and middle lobes of liver were removed. The rats were killed by cervical vertebra dislocation at 0.5, 1, 2, 4, 6, 8, 12, 16, 18, 24, 30, 36, 42, 48, 54, 60, 66, 72, 96, 120, 144 and 168 h after PH and the regenerating livers were observed at corresponding time points. The livers were rinsed three times in PBS at 4°C, then 100-200 mg liver tissue was cut from the middle of right lobe. Six samples were taken from each group and mixed into 1-2 g (0.1-0.2 g × 6) liver tissue, then stored at -80°C. The sham-operation (SO) groups underwent the same PH without removal of the liver lobes. The animal protection laws in China were strictly followed.
Total RNA was isolated from frozen livers according to the manual of Trizol kit (Invittrogen)[18] and then purified based on the guide of RNeasy mini kit (Qiagen)[19]. Total RNA sample was checked to exhibit a 2:1 ratio of 28S to 18S rRNA intensities by agarose electrophoresis (180V, 0.5 h). Total RNA concentration and purity were estimated by optical density measurements at 260/280 nm[20].
As a template, 1-8 μg total RNA was used for cDNA synthesis. cDNA and cRNA synthesis was carried out as previously described[21]. cRNA labeled with biotin was synthesized using 12 μL synthesized cDNA as a template, cDNA and cRNA were purifid[21]. Measurement of concentration, purity and quality of cDNA and cRNA was performed as previously reported[20].
Fifteen microliters (1 μg/μL) cRNA incubated with 5 × fragmentation buffer at 94°C for 35 min was digested into 35-200 bp fragments. Rat genome 230 2.0 microarry produced by Affymetrix was prehybridized, then the hybridization buffer was centrifuged at 60 r/min for 16 h at 45°C. The microarray was washed and stained with GeneChip fluidics station 450 (Affymetrix Inc., USA). The chips were scanned by GeneChip scan 3000 (Affymetrix Inc., USA), and the signal values of gene expression were observed[22].
The normalized signal values, signal detections (P, A, M) and experiment/control (Ri) were obtained by quantifing and normalizing the signal values using GCOS1.2[22].
To minimize errors in the microarray analysis, each analysis was performed three times by rat genome 230 2.0 microarray. Results with a maximal total ratio (Rm) and the average of three housekeeping genes (β-actin, hexokinase and glyseraldehyde-3-phosphate dehydrogenase) approached to 1.0 (Rh) were taken as a reference. Modified data were generated by applying a correction factor (Rm/Rh) multiplying the ratio of every gene in Rh at each time point. To remove spurious gene expression changes resulting from errors in the microarray analysis, the gene expression profiles at 0-4 h, 6-12 h and 12-24 h after PH were reorganized by normalization analysis program (NAP) software according to the cell cycle progression of regenerating hepatocytes. Data statistics and cluster analysis were done using GeneMath, GeneSpring, Microsoft Excel softwares[22-24].
Firstly, the nomenclature of structure and activity of hepatitis virus (e.g. hepatitis B virus) were adopted from the NCBI (http://http://www.ncbi.nlm.nih.gov/) and GENEONTOLOGY database (http://www.geneontology.org/), and input into the databases at AmiGO (http://www.godatabase.org/), NCBI (http://www.ncbi.nlm.nih.gov/), RGD (rgd.mcw. edu/), MGI (http://www.informatics.jax.org/), UniProt (http://www.pir.uniprot.org/) to identify the rat, mouse and human genes associated with hepatitis virus infection. Then the genes associated with hepatitis virus infection were collated. The results of this analysis were codified, and compared with the results obtained for mouse and human searches in order to identify human genes which are different from those of rats. Compared these genes with the analysis output of the Rat Genome 230 2.0 array, those genes which showed a greater than twofold change in expression level, observed as meaningful expression changes[25], were referred to as rat homologous genes or rat specific genes associated with hepatitis virus infection. Genes, which displayed reproducible results with three independent analyses with the chip and showed a greater than twofold change in expression level in at least one time point during liver regeneration with significant difference (0.01 ≤P < 0.05) or extremely significant difference (P≤ 0.01) between PH and SO, were referred to as associated with liver regeneration.
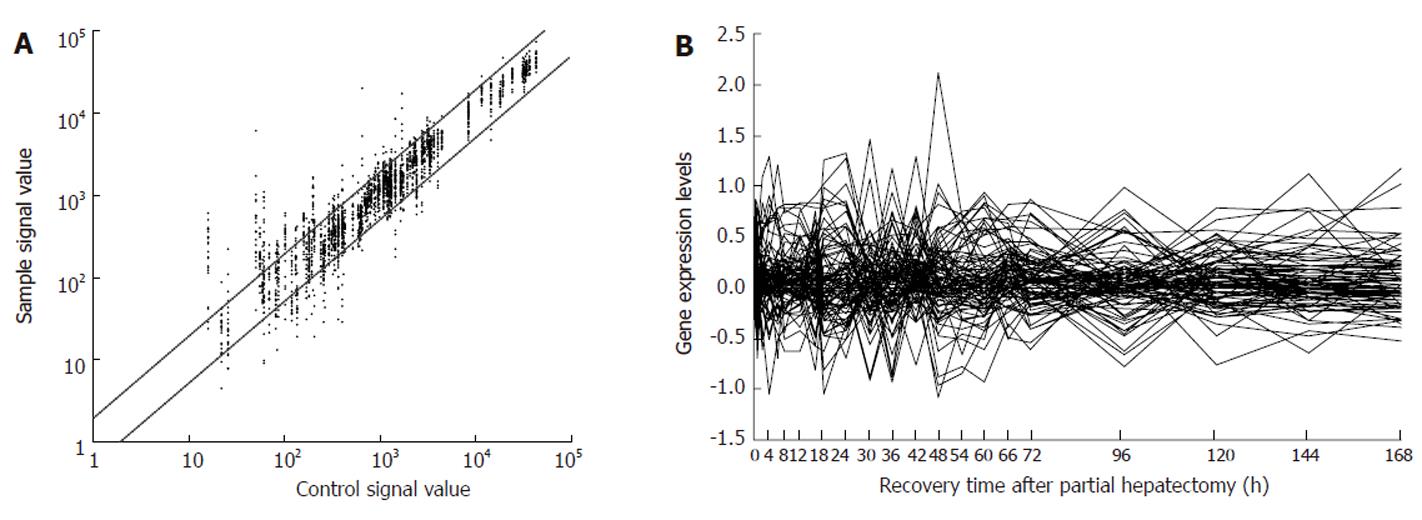
According to the data from AmiGO, NCBI, RGD, MGI and UniProt databases, 204 genes were associated with hepatitis virus infection, and 193 of the above 204 genes were contained in rat genome 230 2.0 array. Among them, 88 genes revealed meaningful changes in expression at least at one time point after partial hepatectomy (PH), and showed significant or extreme significant differences in expression when compared with sham operation (SO), and were reproducible in three detections by rat genome 230 2.0 array, suggesting that the genes were associated with LR (Table 1). The analysis indicated that 37 genes were up regulated, 34 genes down-regulated, and 17 genes up/down-regulated during LR. Total expression frequencies of up- and down-regulated genes were 359 and 149, respectively (Figure 1A). The expression patterns varied with the phases in regenerating liver. At the initiation stage of LR (0.5-4 h after PH), 20 genes displayed up-regulation, 14 genes down-, 2 genes up/down-regulation; at the transition phase from G0 to G1 (4-6 h after PH), 15 genes revealed up-, 11 genes down-regulation; at cell proliferation phase (6-66 h after PH), 40 genes showed up-, 26 genes down-regulation, 12 genes up/down-regulation; at cell differentiation and structure-function reorganization stage (66-168 h after PH), 35 genes displayed up-, 17 genes down-, 5 genes up/down-regulation (Figure 1B).
| Gene Abbr. | Invovled in others | Fold difference | Gene Abbr. | Invovled in others | Fold difference | Gene Abbr. | Invovled in others | Fold difference | Gene Abbr. | Invovled in others | Fold difference |
| 1 Virus life cycle and virion | Ifng | 6.5 | Bcl2 | 0.3 | Rgd1306332 | 0.3 | |||||
| Abce1 | 2.7 | Il1b | 3 | 0.4 | Casp8 | 10.6 | Serpinf2 | 0.2 | |||
| Ccl2 | 3 | 128.0 | Il1r1 | 0.5 | Ccl2 | 128.0 | Snrpd1 | 4.3 | |||
| Ccl4 | 0.2, 3.0 | Il4 | 2.6 | Cyp17a1 | 0.3 | Tap1 | 2.2 | ||||
| Ctbp2 | 1.4 | Ilf3 | 4.0 | Cyp2d6 | 0.3 | Tgfb1 | 4.0 | ||||
| Ctse | 2.0 | Mapk8 | 19.7 | Ddx3x | 0.4 | Tlr2 | 10.6 | ||||
| Gfi1 | 0.2, 2.4 | Mbl2 | 0.2 | Dffa | 0.3, 2.8 | Tnf | 2 | 3.2 | |||
| Hbxip | 2 | 2.0 | Mmp9 | 9.5 | E2f1 | 21.2 | Tp53 | 2 | 2.9 | ||
| Hipk2 | 0.2, 2.8 | Nfkb1 | 2.3 | Eif4a1 | 4.1 | Vapa | 28.8 | ||||
| Hrmt1l2 | 0.5, 2.3 | Nr4a1 | 7.5 | Ephx1 | 0.4, 2.8 | Vipr1 | 2.3 | ||||
| Npap60 | 1.3 | Nrf1 | 2.4 | Hm13 | 0.1 | Wnt1 | 0.5 | ||||
| Oprk1 | 1.6 | Pcna | 10.6 | Ifna1 | 13.0 | Ywhaz | 2.3 | ||||
| Ppia | 2.6 | Ptgs2 | 3 | 2.1 | Ikbkb | 0.3 | 4 Involved in other hepatitis | ||||
| Tnip1 | 0.5 | Ptk2b | 3.6 | Il1b | 2 | 0.4 | virus | ||||
| Ubp1 | 0.5 | Rfx1 | 7.9 | Irf1 | 0.3 | Adarb1 | 0.4 | ||||
| Wwp1 | 3.9 | Serpinb3 | 0.1 | Ltbr | 0.4 | Ambp | 5.1 | ||||
| 2 Hepatitis B virus | Shc1 | 0.5 | Mapk1 | 2.7 | Btbd1 | 0.5 | |||||
| Creb1 | 0.5 | Tcf1 | 6.8 | Mbp | 0.4 | Copg | 0.1 | ||||
| Ddb2 | 2.8 | Timp1 | 8.6 | Nfkbib | 11.8 | Copg2 | 2.8 | ||||
| Egr2 | 6.8 | Timp3 | 0.5 | Nolc1 | 3.7 | Dap3 | 5.5 | ||||
| Esr1 | 6.1 | Tnf | 3 | 3.2 | Pitx1 | 4.6 | Havcr1 | 16.0 | |||
| Foxa2 | 0.4 | Tp53 | 3 | 2.9 | Ptbp2 | 2.2 | Nd1 | 0.4 | |||
| Hbxip | 0.5 | Xpo1 | 3.2 | Pten | 0.5 | Snrpn | 7.3 | ||||
| Hnf4a | 4.5 | 3 Hepatitis C virus | Ptgs2 | 2 | 0.1, 2.1 | Sos1 | 0.4, 14.9 | ||||
| Hspa5 | 0.1 | Apoe | 0.1 | Rb1 | 2.6 |
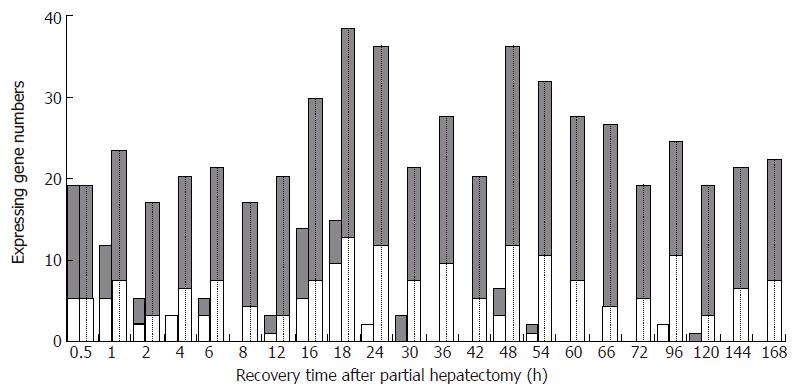
At each time point of LR, the number of initially up-, down-regulated and totally up-, down-regulated genes was 13 and 5 at 0.5 h; 6, 5 and 15, 7 at 1 h; 3, 2 and 13, 3 at 2 h; 0, 3 and 13, 6 at 4 h; 2, 3 and 13, 7 at 6 h; 0, 0 and 12, 4 at 8 h; 2, 1 and 16, 3 at 12 h; 8, 5 and 21, 7 at 16 h; 5, 9 and 24, 12 at 18 h; 0, 2 and 23, 11 at 24 h; 3, 0 and 13, 7 at 30 h; 0, 0 and 17, 9 at 36 h; 0, 0 and 14, 5 at 42 h; 3, 3 and 23, 11 at 48 h; 1, 1 and 20, 10 at 54 h; 0, 0 and 19, 7 at 60 h; 0, 0 and 21, 4 at 66 h; 0, 0 and 13, 5 at 72 h; 0, 2 and 13, 10 at 96 h; 1, 0 and 15, 3 at 120 h; 0, 0 and 14, 6 at 144 h; 0, 0 and 14, 7 at 168 h (Figure 2).
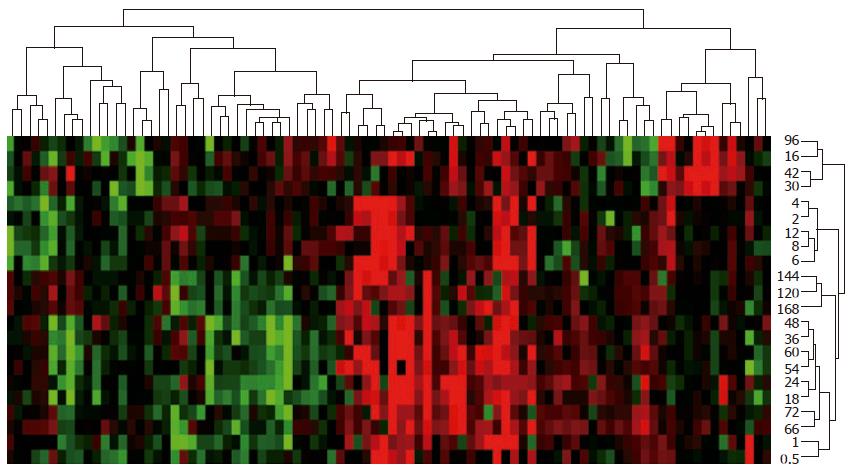
A total of 88 genes during LR could be divided into as following: 37 up-regulated, 9 predominantly up-regulated, 34 down-regulated, 6 predominantly down-regulated, and 2 up/down-regulated genes based on their similarity in expression (Figure 3). These 88 genes during liver regeneration could also be classified based on time relevance into 12 groups (0.5 and 1 h, 2 and 4 h, 6 h, 8 and 12 h, 16 and 96 h, 18 and 24 h, 30 and 42 h, 36 and 48 h, 54 and 60 h, 66 and 72 h, 120 and 144 h, 168 h) in which the number of up- and down-regulated genes was 28 and 12, 26 and 9, 13 and 7, 28 and 7, 34 and 17, 47 and 23, 27 and 12, 40 and 20, 39 and 17, 34 and 9, 29 and 9, 14 and 7 (Figure 3).
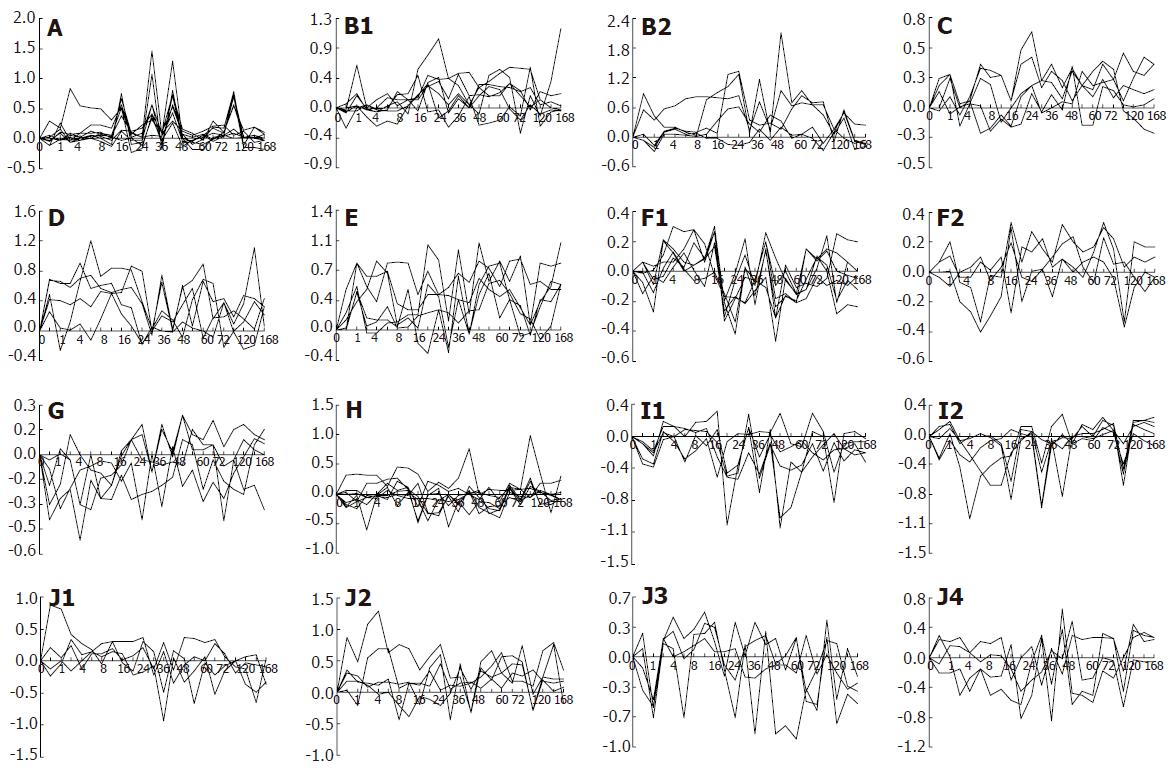
The expression patterns of 88 genes during liver regeneration might be categorized into 23 types according to the expression changes: (1) up-regulated expression at one time point, i.e. 30, 48, 120 h after PH (Figure 4A-C and E) with 4 genes involved; (2) up-regulated expression at two time points, i.e. 30 and 42 h, 16 and 42 h, 16 and 96 h, 18 and 54 h (Figure 4A and B) with 4 genes involved; (3) up-regulated expression at multiple time points (Figure 4A and B) with 5 genes involved; (4) up-regulated expression at one phase, i.e. 1-168 h, 16-96 h (Figure 4B and E) with 2 genes involved; (5) up-regulation expression at two phases, i.e. 16-24 and 42-48 h (Figure 4B) with 1 gene involved; (6) up-regulated expression at multiple phases (Figure 4E) with 1 gene involved; (7) up-regulated expression at one time point/one phase, i.e. 54 and 144-168 h (Figure 4C) with 1 gene involved; (8) up-regulated expression at one time point/two phases (Figure 4B-D) with 4 genes involved; (9) up-regulated expression at one time point/multiple phases (Figure 4E and J) with 2 genes involved; (10) up-regulated expression at two time points/one phase (Figure 4B, D and E) with 3 genes involved; (11) up-regulated expression at two time points/two phases (Figure 4B and D) with 2 genes involved; (12) up-regulated expression at two time points/multiple phases (Figure 4B, D and E) with 3 genes involved; (13) up-regulated expression at multiple time points/one phase (Figure 4A and C) with 2 genes involved; (14) up-regulated expression at multiple time points/two phases (Figure 4C, D and E) with 3 genes involved; (15) down-regulated expression at one time point, i.e. 2, 4, 6, 16, 18, 24, 48 , 96h (Figure 4F, G and H) with 14 genes involved; (16) down-regulated expression at two time points, i.e. 0.5 and 4 h, 16 and 96 h, 18 and 54 h (Figure 4G, H and I) with 3 genes involved; (17) down-regulated expression at multiple time points (Figure 4G, H and I) with 6 genes involved; (18) down-regulated expression at one phase, i.e. 4-6 h, 18-24 h, 54-60 h (Figure 4G and H) with 3 genes involved; (19) down-regulated expression at one time point and one phase, i.e. 1 and 144-168 h, 48 and 18-24 h, 30 and 2-8 h (Figure 4H, I and J) with 3 genes involved; (20) down-regulated expression at one time point/two phases (Figure 4I) with 1 gene involved; (21) down-regulated expression at two time points/one phase (Figure 4I) with 1 gene involved; (22) down-regulated expression at two time points/two phases (Figure 4I) with 3 genes involved; (23) up/down-regulated expression (Figure 4B, H and J) with 17 genes involved.
The role of 193 genes associated with hepatitis virus infection in liver regeneration was studied in this study. Among the 45 genes associated with hepatitis virus life cycle and virion, peptidylprolyl isomerase A (PPIA), nuclear pore associated protein (NPAP60) and opioid receptor kappa 1 (OPRκ1) are involved in viral infection[26]. Eight genes, including chemokine ligand 2 (CCL2), WW domain containing E3 ubiquitin protein ligase 1 (WWP1), chemokine ligand 3 (CCL3), C-terminal binding protein 2 (CTBP2), cathepsin E (CTSE), TNFAIP3 interacting protein 1 (TNIP1), upstream binding protein 1 (UBP1) and heterogeneous nuclear ribonucleoproteins methyltransferase-like 2 (HRMT1L2), participate in the viral genome replication and transcription[27,28]. Hbx interacting protein (HBXIP) suppresses HBV X protein activity[29]. ATP-binding cassette sub-family E member 1 (ABCE1) plays a part in viral capsid assembly[30]. Homeodomain interacting protein kinase 2 (HIPK2) relates to induction of apoptosis[31]. Growth factor independent 1 (GFI1) controls the differentiation of dendritic cells into macrophages[32]. The above genes tend to have a same or similar expression at some time points, but a different expression at other time points which may enhance inflammatory reaction and immunity of regenerating liver.
Of the 65 genes associated with hepatitis B virus infection, forkhead box A2 (FOXA2), interleukin 4 (IL4) and transcription factor 1 (TCF1) may restrain HBV gene replication and RNA transcription[33,34], whereas regulatory factor X 1 (RFX1) promotes RNA transcription by binding to HBV enhancer[35]. Tissue inhibitor of metalloproteinases 1, 3 (TIMP1, TIMP3) exhibits low level expression after HBV transinfection[36], but matrix metallopeptidase 9 (MMP9) displays high activity after HBV transinfection[36]. Interferon gamma (IFNγ) supresses hepatic cell proliferation[37]. CAMP responsive element binding protein 1 (CREB1), nuclear respiratory factor 1 (NRF1) and hepatocyte nuclear factor 4 alpha (HNF4α) activate HBV promoter[34,38,39]. Nine genes, such as early growth response 2 (EGR2), proliferating cell nuclear antigen (PCNA), tumor suppressor p53 (TP53), serpin peptidase inhibitor clade B member 3 (SERPINB3), prostaglandin-endoperoxide synthase 2 (PTGS2), SHC transforming protein 1 (SHC1), interleukin enhancer binding factor 3 (ILF3), nuclear factor of kappa light chain gene enhancer in B-cells 1 (NFκB1) and mitogen-activated protein kinase 8 (MAPK8), accelerate cell proliferation[40-45]. Tumor necrosis factor (TNF) and estrogen receptor 1 (ESR1) increase cell susceptibility to HBV infection[46,47]. 70kDa heat shock protein 5 (HSPA5) plays a role in anti-apoptosis[48]. Protein tyrosine kinase 2 beta (PTK2β) promotes migration and invasion of glioblastoma[49]. Exportin 1 (XPO1) takes part in HBV-induced aberrant centriole replication and abnormal mitotic spindles[50]. Mannose binding lectin 2 (MBL2) participates in HBV infection[51]. Interleukin 1 beta (IL1β) may enhance resistance to chronic diseases[52]. Interleukin 1 receptor type I (IL1R1) elevates blood-brain barrier[53]. Damage specific DNA binding protein 2 (DDB2) promotes turnover of HBV X protein[54]. Nuclear receptor subfamily 4 group A member 1 (NR4A1) enhances role of the HBx-induced Fas/FasL signaling pathway[55]. The above genes may have same or similar expression changes at some time points, but different expression changes at other time points, suggesting that they promote inflammation recovery and strengthen anti-hepatitis B virus infection ability.
Among the 90 genes associated with hepatitis C virus infection, Three genes concluding polypyrimidine-tract binding protein 2 (PTBP2), mitogen-activated protein kinase 1 (MAPK1) and interferon alpha 1 (IFNα1) play a role in suppression of hepatitis C virus replication[56-58]. Nucleolar and coiled-body phosphoprotein 1 (NOLC1), transporter 1 ATP-binding cassette sub-family B (TAP1) and eukaryotic translation initiation factor 4A1 (EIF4A1) promote protein biosynthesis[59]. Six genes including toll-like receptor (TLR2), transforming growth factor beta 1 (TGFβ1), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAζ), vasoactive intestinal peptide receptor 1 (VIPR1), cytochrome P450 family 2 subfamily D polypeptide 6 (CYP2D6) and lymphotoxin B receptor (LTBR), are related to immune reponse[60,61]. VAMP-associated protein A (VAPA) accelerates HCV replication[62]. Nuclear factor of kappa light chain gene enhancer in B-cell inhibitor beta (NFκBIβ) and E2F transcription factor 1 (E2F1) have transcriptional activator activity[63]. Inhibitor of kappaB kinase beta (IKκBKβ) inhibits activation of NFκB. Small nuclear ribonucleoprotein D1 (SNRPD1) and serine peptidase inhibitor clade F member 2 (SERPINF2) can repair liver damage[64,65]. Paired-like homeodomain transcription factor 1 (PITX1) modulates interferon expression[66]. Genetic polymorphisms of cytochrome P450 family 17 subfamily alpha polypeptide 1 (CYP17α1) and epoxide hydrolase 1 (EPHX1) are closely associated with liver disease[67,68]. Four genes containing interferon regulatory factor 1 (IRF1), phosphatase and tensin homolog (PTEN), myelin basic protein (MBP) and DEAD box polypeptide 3 X-linked (DDX3X) are associated with regulation of liver disease[69-72]. Apolipoprotein E (APOE) has a role in liver protection[73]. Retinoblastoma 1 (RB1) and caspase 8 (CASP8) inhibit cell growth[74,75]. Wingless-type MMTV integration site family member 1 (WNT1) can inhibit cell apoptosis[76]. B-cell leukemia/lymphoma 2 (BCL2) is associated with infectious vasculitis[77]. Histocompatibility antigen 13 (HM13) is responsible for immunological recognition[78]. HM13 is highly expressed in human liver HL-7702 cells as HCV NS3-transactivated protein 1 (RGD1306332)[79]. DNA fragmentation factor alpha subunit (DFFα) plays a role in suppressing tumors[80]. The above genes may have the same or similar expression changes at some time points, different expression changes at other time points, thus promoting inflammatory reaction and anti-infection ability of regenerating liver.
In summary, the expression changes of hepatitis virus infection-associated genes after rat partial hepatectomy can be analyzed with high-throughput gene expression profiling. Immunological competence, abilities of anti-inflammation and anti-infection are increased during liver regeneration. Rat genome 230 2.0 array is a useful tool for analyzing hepatitis virus infection and liver disease at transcriptional level. However, DNA→ mRNA→protein is affected by many factors including protein interaction. Further study is needed to confirm the results at cell level.
S- Editor Liu Y L- Editor Wang XL E- Editor Bi L
| 1. | Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1091] [Cited by in RCA: 1175] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 2. | Michalopoulos GK, DeFrances M. Liver regeneration. Adv Biochem Eng Biotechnol. 2005;93:101-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 3. | Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45-S53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1126] [Cited by in RCA: 1204] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 4. | Xu CS, Zhao LF, Yang KJ, Zhang JB. [The origination and action of the hepatic stems cells]. Shi Yan Sheng Wu Xue Bao. 2004;37:72-77. [PubMed] |
| 5. | Xu CS, Chang CF, Yuan JY, Li WQ, Han HP, Yang KJ, Zhao LF, Li YC, Zhang HY, Rahman S. Expressed genes in regenerating rat liver after partial hepatectomy. World J Gastroenterol. 2005;11:2932-2940. [PubMed] |
| 6. | Yap SF. Hepatitis B: review of development from the discovery of the "Australia Antigen" to end of the twentieth Century. Malays J Pathol. 2004;26:1-12. [PubMed] |
| 7. | Li H, Wang L, Wang SS, Gong J, Zeng XJ, Li RC, Nong Y, Huang YK, Chen XR, Huang ZN. Research on optimal immunization strategies for hepatitis B in different endemic areas in China. World J Gastroenterol. 2000;6:392-394. [PubMed] |
| 8. | Merle P. [Epidemiology, natural history and pathogenesis of hepatocellular carcinoma]. Cancer Radiother. 2005;9:452-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Leung N. HBV and liver cancer. Med J Malaysia. 2005;60 Suppl B:63-66. [PubMed] |
| 10. | Ocama P, Opio CK, Lee WM. Hepatitis B virus infection: current status. Am J Med. 2005;118:1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Pujol FH, Devesa M. Genotypic variability of hepatitis viruses associated with chronic infection and the development of hepatocellular carcinoma. J Clin Gastroenterol. 2005;39:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Iacono KT, Kazi L, Weiss SR. Both spike and background genes contribute to murine coronavirus neurovirulence. J Virol. 2006;80:6834-6843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Lefkowitch JH. Hepatobiliary pathology. Curr Opin Gastroenterol. 2006;22:198-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Lai HS, Chen Y, Lin WH, Chen CN, Wu HC, Chang CJ, Lee PH, Chang KJ, Chen WJ. Quantitative gene expression analysis by cDNA microarray during liver regeneration after partial hepatectomy in rats. Surg Today. 2005;35:396-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1089] [Cited by in RCA: 1126] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 16. | Schadt EE, Edwards SW, GuhaThakurta D, Holder D, Ying L, Svetnik V, Leonardson A, Hart KW, Russell A, Li G. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 2004;5:R73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Higgins GM, Anderson RM. Experimental pathology of the liver: restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186-202. |
| 18. | Knepp JH, Geahr MA, Forman MS, Valsamakis A. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J Clin Microbiol. 2003;41:3532-3536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Nuyts S, Van Mellaert L, Lambin P, Anné J. Efficient isolation of total RNA from Clostridium without DNA contamination. J Microbiol Methods. 2001;44:235-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633-1648. [PubMed] |
| 21. | Li L, Roden J, Shapiro BE, Wold BJ, Bhatia S, Forman SJ, Bhatia R. Reproducibility, fidelity, and discriminant validity of mRNA amplification for microarray analysis from primary hematopoietic cells. J Mol Diagn. 2005;7:48-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Hood L. Leroy Hood expounds the principles, practice and future of systems biology. Drug Discov Today. 2003;8:436-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863-14868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12067] [Cited by in RCA: 10557] [Article Influence: 391.0] [Reference Citation Analysis (0)] |
| 24. | Werner T. Cluster analysis and promoter modelling as bioinformatics tools for the identification of target genes from expression array data. Pharmacogenomics. 2001;2:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Valverde O, Mantamadiotis T, Torrecilla M, Ugedo L, Pineda J, Bleckmann S, Gass P, Kretz O, Mitchell JM, Schütz G. Modulation of anxiety-like behavior and morphine dependence in CREB-deficient mice. Neuropsychopharmacology. 2004;29:1122-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 93] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 26. | Sokolskaja E, Berthoux L, Luban J. Cyclophilin A and TRIM5alpha independently regulate human immunodeficiency virus type 1 infectivity in human cells. J Virol. 2006;80:2855-2862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 27. | Henningsson F, Yamamoto K, Saftig P, Reinheckel T, Peters C, Knight SD, Pejler G. A role for cathepsin E in the processing of mast-cell carboxypeptidase A. J Cell Sci. 2005;118:2035-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Araya N, Hiraga H, Kako K, Arao Y, Kato S, Fukamizu A. Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem Biophys Res Commun. 2005;329:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22:2729-2740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 353] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 30. | Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE,Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L,Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G,di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR,Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M,Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D,Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G,Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schönbach C, Sekiguchi K,Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K,Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S,Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y,Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S,Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science. 2005;309:1559-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2883] [Cited by in RCA: 2677] [Article Influence: 133.9] [Reference Citation Analysis (0)] |
| 31. | Di Stefano V, Blandino G, Sacchi A, Soddu S, D'Orazi G. HIPK2 neutralizes MDM2 inhibition rescuing p53 transcriptional activity and apoptotic function. Oncogene. 2004;23:5185-5192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Rathinam C, Geffers R, Yücel R, Buer J, Welte K, Möröy T, Klein C. The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity. 2005;22:717-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 97] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 33. | Lin SJ, Shu PY, Chang C, Ng AK, Hu CP. IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. J Immunol. 2003;171:4708-4716. [PubMed] |
| 34. | Zheng Y, Li J, Ou JH. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J Virol. 2004;78:6908-6914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 86] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 35. | Katan-Khaykovich Y, Shaul Y. Nuclear import and DNA-binding activity of RFX1. Evidence for an autoinhibitory mechanism. Eur J Biochem. 2001;268:3108-3116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 36. | Kim JR, Kim CH. Association of a high activity of matrix metalloproteinase-9 to low levels of tissue inhibitors of metalloproteinase-1 and -3 in human hepatitis B-viral hepatoma cells. Int J Biochem Cell Biol. 2004;36:2293-2306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Shen H, Zhang M, Minuk GY, Gong Y. Different effects of rat interferon alpha, beta and gamma on rat hepatic stellate cell proliferation and activation. BMC Cell Biol. 2002;3:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 38. | Tacke F, Liedtke C, Bocklage S, Manns MP, Trautwein C. CREB/PKA sensitive signalling pathways activate and maintain expression levels of the hepatitis B virus pre-S2/S promoter. Gut. 2005;54:1309-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Tokusumi Y, Zhou S, Takada S. Nuclear respiratory factor 1 plays an essential role in transcriptional initiation from the hepatitis B virus x gene promoter. J Virol. 2004;78:10856-10864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Yoo YG, Lee MO. Hepatitis B virus X protein induces expression of Fas ligand gene through enhancing transcriptional activity of early growth response factor. J Biol Chem. 2004;279:36242-36249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 41. | Shen LJ, Zhang HX, Zhang ZJ, Li JY, Chen MQ, Yang WB, Huang R. Detection of HBV, PCNA and GST-pi in hepatocellular carcinoma and chronic liver diseases. World J Gastroenterol. 2003;9:459-462. [PubMed] |
| 42. | Qu JH, Zhu MH, Lin J, Ni CR, Li FM, Zhu Z, Yu GZ. Effects of hepatitis B virus on p53 expression in hepatoma cell line SMMU-7721. World J Gastroenterol. 2005;11:6212-6215. [PubMed] |
| 43. | Ruvoletto MG, Tono N, Carollo D, Vilei T, Trentin L, Muraca M, Marino M, Gatta A, Fassina G, Pontisso P. Surface expression of squamous cell carcinoma antigen (SCCA) can be increased by the preS1(21-47) sequence of hepatitis B virus. J Gen Virol. 2004;85:621-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 44. | Carloni V, Mazzocca A, Ravichandran KS. Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene. 2004;23:1566-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 45. | Yang L, Magness ST, Bataller R, Rippe RA, Brenner DA. NF-kappaB activation in Kupffer cells after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2005;289:G530-G538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 46. | Suneetha PV, Sarin SK, Goyal A, Kumar GT, Shukla DK, Hissar S. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 47. | Deng G, Zhou G, Zhai Y, Li S, Li X, Li Y, Zhang R, Yao Z, Shen Y, Qiang B. Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology. 2004;40:318-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 48. | Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915-20924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 575] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 49. | Lipinski CA, Tran NL, Menashi E, Rohl C, Kloss J, Bay RC, Berens ME, Loftus JC. The tyrosine kinase pyk2 promotes migration and invasion of glioma cells. Neoplasia. 2005;7:435-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 50. | Forgues M, Difilippantonio MJ, Linke SP, Ried T, Nagashima K, Feden J, Valerie K, Fukasawa K, Wang XW. Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol Cell Biol. 2003;23:5282-5292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 51. | Song le H, Binh VQ, Duy DN, Jüliger S, Bock TC, Luty AJ, Kremsner PG, Kun JF. Mannose-binding lectin gene polymorphisms and hepatitis B virus infection in Vietnamese patients. Mutat Res. 2003;522:119-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 52. | Nickerson M, Elphick GF, Campisi J, Greenwood BN, Fleshner M. Physical activity alters the brain Hsp72 and IL-1beta responses to peripheral E. coli challenge. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1665-R1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 53. | Konsman JP, Vigues S, Mackerlova L, Bristow A, Blomqvist A. Rat brain vascular distribution of interleukin-1 type-1 receptor immunoreactivity: relationship to patterns of inducible cyclooxygenase expression by peripheral inflammatory stimuli. J Comp Neurol. 2004;472:113-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 185] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 54. | Bergametti F, Sitterlin D, Transy C. Turnover of hepatitis B virus X protein is regulated by damaged DNA-binding complex. J Virol. 2002;76:6495-6501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 55. | Lee MO, Kang HJ, Cho H, Shin EC, Park JH, Kim SJ. Hepatitis B virus X protein induced expression of the Nur77 gene. Biochem Biophys Res Commun. 2001;288:1162-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 56. | Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology. 2005;335:72-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 69] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 57. | Ishida H, Ohkawa K, Hosui A, Hiramatsu N, Kanto T, Ueda K, Takehara T, Hayashi N. Involvement of p38 signaling pathway in interferon-alpha-mediated antiviral activity toward hepatitis C virus. Biochem Biophys Res Commun. 2004;321:722-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 58. | Zhang T, Lin RT, Li Y, Douglas SD, Maxcey C, Ho C, Lai JP, Wang YJ, Wan Q, Ho WZ. Hepatitis C virus inhibits intracellular interferon alpha expression in human hepatic cell lines. Hepatology. 2005;42:819-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 59. | Goggin MM, Nelsen CJ, Kimball SR, Jefferson LS, Morley SJ, Albrecht JH. Rapamycin-sensitive induction of eukaryotic initiation factor 4F in regenerating mouse liver. Hepatology. 2004;40:537-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 60. | Seki E, Tsutsui H, Iimuro Y, Naka T, Son G, Akira S, Kishimoto T, Nakanishi K, Fujimoto J. Contribution of Toll-like receptor/myeloid differentiation factor 88 signaling to murine liver regeneration. Hepatology. 2005;41:443-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 128] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 61. | Anand S, Wang P, Yoshimura K, Choi IH, Hilliard A, Chen YH, Wang CR, Schulick R, Flies AS, Flies DB. Essential role of TNF family molecule LIGHT as a cytokine in the pathogenesis of hepatitis. J Clin Invest. 2006;116:1045-1051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 62. | Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480-3488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 265] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 63. | de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem. 2003;278:42041-42049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 166] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 64. | Iwai A, Hasumura Y, Nojima T, Takegami T. Hepatitis C virus nonstructural protein NS3 binds to Sm-D1, a small nuclear ribonucleoprotein associated with autoimmune disease. Microbiol Immunol. 2003;47:601-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 65. | Okada K, Ueshima S, Imano M, Kataoka K, Matsuo O. The regulation of liver regeneration by the plasmin/alpha 2-antiplasmin system. J Hepatol. 2004;40:110-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 66. | Ghosh AK, Majumder M, Steele R, Ray R, Ray RB. Modulation of interferon expression by hepatitis C virus NS5A protein and human homeodomain protein PTX1. Virology. 2003;306:51-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 67. | Rossi L, Leveri M, Gritti C, De Silvestri A, Zavaglia C, Sonzogni L, Silvestri L, Civardi E, Mondelli MU, Silini EM. Genetic polymorphisms of steroid hormone metabolizing enzymes and risk of liver cancer in hepatitis C-infected patients. J Hepatol. 2003;39:564-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 68. | Sonzogni L, Silvestri L, De Silvestri A, Gritti C, Foti L, Zavaglia C, Bottelli R, Mondelli MU, Civardi E, Silini EM. Polymorphisms of microsomal epoxide hydrolase gene and severity of HCV-related liver disease. Hepatology. 2002;36:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 69. | Saito H, Tada S, Wakabayashi K, Nakamoto N, Takahashi M, Nakamura M, Ebinuma H, Ishii H. The detection of IRF-1 promoter polymorphisms and their possible contribution to T helper 1 response in chronic hepatitis C. J Interferon Cytokine Res. 2002;22:693-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 70. | Rahman MA, Kyriazanos ID, Ono T, Yamanoi A, Kohno H, Tsuchiya M, Nagasue N. Impact of PTEN expression on the outcome of hepatitis C virus-positive cirrhotic hepatocellular carcinoma patients: possible relationship with COX II and inducible nitric oxide synthase. Int J Cancer. 2002;100:152-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 71. | Medveczky P, Antal J, Patthy A, Kékesi K, Juhász G, Szilágyi L, Gráf L. Myelin basic protein, an autoantigen in multiple sclerosis, is selectively processed by human trypsin 4. FEBS Lett. 2006;580:545-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 72. | Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, Wu Lee YH. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene. 2006;25:1991-2003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 111] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 73. | Wozniak MA, Itzhaki RF, Faragher EB, James MW, Ryder SD, Irving WL. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology. 2002;36:456-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 132] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 74. | Hassan M, Ghozlan H, Abdel-Kader O. Activation of RB/E2F signaling pathway is required for the modulation of hepatitis C virus core protein-induced cell growth in liver and non-liver cells. Cell Signal. 2004;16:1375-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 75. | Kovalovich K, Li W, DeAngelis R, Greenbaum LE, Ciliberto G, Taub R. Interleukin-6 protects against Fas-mediated death by establishing a critical level of anti-apoptotic hepatic proteins FLIP, Bcl-2, and Bcl-xL. J Biol Chem. 2001;276:26605-26613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 229] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 76. | Almeida M, Han L, Bellido T, Manolagas SC, Kousteni S. Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by beta-catenin-dependent and -independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol Chem. 2005;280:41342-41351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 330] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 77. | Casato M, Mecucci C, Agnello V, Fiorilli M, Knight GB, Matteucci C, Gao L, Kay J. Regression of lymphoproliferative disorder after treatment for hepatitis C virus infection in a patient with partial trisomy 3, Bcl-2 overexpression, and type II cryoglobulinemia. Blood. 2002;99:2259-2261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 78. | Okamoto K, Moriishi K, Miyamura T, Matsuura Y. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J Virol. 2004;78:6370-6380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 79. | Li JW, Li K, Jiang J, Xu XL, Huang ZQ. Construction of eukaryotic expression plasmid containing HCV NS3 segment and protein expression in human HL-7702 hepatocytes. World J Gastroenterol. 2006;12:1038-1042. [PubMed] |
| 80. | Yan B, Wang H, Peng Y, Hu Y, Wang H, Zhang X, Chen Q, Bedford JS, Dewhirst MW, Li CY. A unique role of the DNA fragmentation factor in maintaining genomic stability. Proc Natl Acad Sci USA. 2006;103:1504-1509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |