Published online Nov 21, 2006. doi: 10.3748/wjg.v12.i43.6949
Revised: September 5, 2006
Accepted: September 11, 2006
Published online: November 21, 2006
AIM: To study the differential gene expression profiles of target cells in primary gastric cancer and its metastatic lymph nodes using laser microdissection (LMD) in combination with cDNA microarray.
METHODS: Normal gastric tissue samples from 30 healthy individuals, 36 cancer tissue samples from primary gastric carcinoma and lymph node metastasis tissue samples from 58 patients during gastric cancer resection were obtained using LMD in combination with cDNA microarray independently. After P27-based amplification, aRNA from 36 of 58 patients (group 1) with lymph node metastasis and metastatic tissue specimens from the remaining 22 patients (group 2) were applied to cDNA microarray. Semiquantitative reverse transcriptase polymerase chain reaction (RT-PCR) and immunohistochemical assay verified the results of microarray in group 2 and further identified genes differentially expressed in the progression of gastric cancer.
RESULTS: The expression of 10 genes was up-regulated while the expression of 15 genes was down-regulated in 22 gastric carcinoma samples compared with that of genes in the normal controls. The results were confirmed at the level of mRNA and protein, and suggested that four genes (OPCML, RNASE1, YES1 and ACK1) could play a key role in the tumorigenesis and metastasis of gastric cancer. The expression pattern of 3 genes (OPCML, RNASE1 and YES1) was similar to tumor suppressor genes. For example, the expression level of these genes was the highest in normal gastric epithelium, which was decreased in primary carcinoma, and further decreased in metastatic lymph nodes. On the contrary, the expression pattern of gene ACK1 was similar to that of oncogene. Four genes were further identified as differentially expressed genes in the majority of the cases in the progression of gastric cancer.
CONCLUSION: LMD in combination with cDNA microarray provides a unique support foe the identification of early expression profiles of differential genes and the expression pattern of 3 genes (OPCML, RNASE1 and YES1) associated with the progression of gastric cancer. Further study is needed to reveal the molecular mechanism of lymph node metastasis in patients with gastric cancer.
- Citation: Wang L, Zhu JS, Song MQ, Chen GQ, Chen JL. Comparison of gene expression profiles between primary tumor and metastatic lesions in gastric cancer patients using laser microdissection and cDNA microarray. World J Gastroenterol 2006; 12(43): 6949-6954
- URL: https://www.wjgnet.com/1007-9327/full/v12/i43/6949.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i43.6949
Gastric cancer is one of the leading causes of cancer death in the world, its clinical behavior depends on the potential metastasis of the tumor, and the prognosis of advanced gastric cancers remains very poor. Until now, several molecules have been reported to play an important role in gastrointestinal tumorigenesis and tumor metastasis[1-3], but the molecular mechanisms involved in tumor development and progression remain unclear in gastric cancer[1-3].
In this study, using the combined methods of laser microdissection (LMD), P27-based RNA amplification, and cDNA microarray, we evaluated the differentially expressed genes in primary carcinoma cells and lymph node metastatic cells in 36 of 58 patients. Moreover, we further identified four differentially expressed genes in the remaining 22 patients with progression of gastric cancer by semiquantitative reverse transcriptase polymerase chain reaction (RT-PCR), and the expression patterns of these four genes were similar to those of tumor suppressor genes or oncogenes.
It has been widely accepted that many malignant tumors contain heterogeneous subpopulations of cells. This heterogeneity exhibits a wide range of genetic, biochemical and immunologic characteristics. It is likely that specific tumor cells or colonies within larger heterogeneous tumor specimens are the forerunners of distant metastases[4]. Therefore, many biologic differences exist in tumor cells of primary carcinoma and metastatic lesions. Furthermore, interaction of tumor cells with their living environment may add more differences to these tumor cells[5]. As a result, tumor metastasis-related genes can be identified by comparing their gene expression profiles.
LMD and cDNA microarray are two of the new emerging techniques in the post-genomic era. LMD is an innovative technique which offers researchers a simple, reliable, rapid and accurate tool for precise and contamination-free procurement of cell groups from tissue sections under direct visualization[6,7].
Large-scale analysis of gene expression with cDNA microarray allows us to evaluate the gene-expression profiles of hundreds to tens of thousands of genes in a single experiment[8]. This techniques is a powerful tool for analyzing the expression of genes which may be correlated with pathological phenotypes of various diseases. However, the expression profile of a specific cell type may be primarily masked or even lost because of the bulky surrounding cells. Therefore, LMD in combination with cDNA microarray can provide a unique opportunity to study gene expression of subpopulations of cells in their native tissue environment in vivo.
Fifty-eight advanced gastric adenocarcinoma (TNM stage III-IV) patients with lymph node metastasis diagnosed by postoperative pathology were investigated in this study. There were 30 male and 28 female patients, whose ages ranged from 45 to 68 years with an average age of 58.7 ± 3.46 years (Table 1). Histologically, 38 patients had moderately differentiated adenocarcinoma and 20 had poorly differentiated adenocarcinoma. All patients underwent gastrectomy with regional lymph nodes dissected and informed consent to participate in this study was obtained from each patient. Tissue blocks of normal gastric epithelium (> 5 cm away from the edge of the tumor), primary tumors and corresponding metastatic lymph nodes were obtained within 30 min after removal from the patient. Each block was cut into 2 pieces, one for routine pathologic diagnosis, and the other for molecular analysis. The latter samples were frozen in liquid nitrogen immediately and stored at -260°C until use.
| Characteristics | n (%) |
| Age (yr) | |
| Range | 48-68 |
| mean ± SD | 58.7 ± 3.46 |
| Median | 59 |
| Mode | 59 |
| Gender | |
| Male | 30 (52) |
| Female | 28 (48) |
| Tumor location | |
| Gastric antrum | 38 (66) |
| Gastric body | 10 (17) |
| Gastric cardia | 10 (17) |
| Anemia | |
| Yes | 48 (83) |
| No | 10 (17) |
| Abdominal pain | |
| Yes | 27 (47) |
| No | 31 (53) |
| Weight los | |
| Yes | 34 (59) |
| No | 24 (41) |
| Occult blood test | |
| Positive | 43 (74) |
| Negative | 25 (26) |
Before sectioning, tissue blocks were embedded in Tissue Tek OCT compound medium (VWR Scientific Products, San Diego, CA, USA) in a cryostat. Then serial 8-micron thick sections were prepared and mounted onto a foiled slide and stored at -70°C until use.
Frozen section slides were stained just before laser microdissection on ice. Briefly, the slides were fixed in 70% ethanol for 30 s and stained with 0.1% toluidine blue (TBO) for 15 s, followed by dehydration in 75%, 95% and 100% ethanol respectively for 5 s and dehydration in xylene for 5 min. Once airdried, the sections were laser microdissected with a LMD system (Leica Microsystems, Wetzlar, Germany) and the target cells were selectively collected. Next, total RNA was extracted from the interest cells independently with the RNA-lyase Micro kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The integrity of the total RNA was measured by Lab-on-chip (Agilent, Palo Alto, USA).
We carried out cDNA microarray analysis in 58 metastatic gastric cancer tissue samples of lymph node metastatic gastric cancer, 36 tissue samples of primary gastric cancer and 30 tissue samples of normal gastric epithelium, all technical services were provided by Shanghai Biochip Corporation (Shanghai, China). Two hundred ng of total RNA was amplified for array analysis, 100 ng of total RNA was not amplified for later certification. After P27-based RNA amplification, aliquots (2.5-microgram) of aRNA from primary carcinoma tissues and their corresponding metastatic lymph nodes were labeled with Cy3-dCTP and Cy5-dCTP, respectively. The labeled probes were hybridized with Human cDNA Chip version 2.0 (SBC-R-HC-100-20, Shanghai, China) containing 13 824 genes (including 10 positive controls and 6 negative ones), and the signals were detected by the Agilent scanner (Agilent, Palo Alto, USA). We set the cutoff values for signal intensities, i.e., the signal to noise ratios of Cy3 or Cy5 must be greater than 2. Genes with Cy3:Cy5 ratios > 4 or < 0.25 in the remaining 22 patients were defined as up- or down-regulated genes.
Nonamplified total RNA (100 ng) was reverse transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen, USA) at 42°C for 60 min and at 70°C for 15 min. Each single-stranded cDNA was diluted for subsequent PCR amplification and the content of cDNA was semiquantitatively normalized by housekeeping gene β-actin. PCR conditions for different genes included an initial denaturation at 94°C for 3 min, followed by 30-35 cycles of denaturation at 94°C for 30 s, annealing at 94°C for 30 s, and elongation at 72°C for 1 min (The primer sequences, annealing temperatures and cycles of each gene are available on request). Amplified PCR products were visualized by electrophoresis on 1% agarose gel containing ethidium bromide.
The slides were incubated with rabbit anti-OPCML antibody (BOSTER, USA) at 1:50 dilution. The sections were then stained with KIT SA1028 (BOSTER, USA) according to the manufacturer’s instructions. The tissue was counterstained with hematoxylin.
Target cells in each sample were successfully laser microdissected. Consequently, about 6 × 106-8 × 106 cells were collected for total RNA extraction, 200-400 ng total RNA was obtained. The integrity of each sample was proven by Lab-on-chip.
All data were expressed as mean ± SD. Statistical significance between the two groups was determined by Student’s t-test using GraphPad Prism version 3.02 for Windows (GraphPad Software Inc., San Diego, USA). P < 0.05 was considered statistically significant.
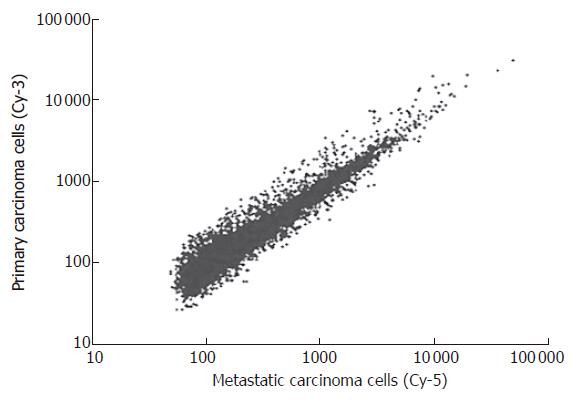
After P27-based amplification, we evaluated the expression profiles of tumor cells of primary gastric cancer and their corresponding metastatic lymph nodes in 36 patients. Scatter plots of cDNA microarray analysis are shown in Figure 1. Amplified aRNA from primary gastric carcinoma cells (Cy3) and metastatic carcinoma cells (Cy5) were labeled and hybridized to the cDNA microarray.
Analysis of the cDNA microarray data showed that 49 genes (including 31 with unknown function) were over-expressed (Cy3:Cy5 > 4) in primary carcinoma cells from 36 patients. On the other hand, 37 genes (including 9 with unknown function) were significantly suppressed (Cy3:Cy5 < 0.25) in primary carcinoma cells from 36 patients. The up-regulated genes were related to cell adhesion, cytoskeleton, cell defense and metabolism. Meanwhile, the down-regulated genes included those associated with cell development, cell cycle, signal transduction, adhesion, cell defense, gene expression and cell metabolism (Table 2).
| GeneBank | Description | Cy3:Cy52 |
| NM_006500 | MCAM (melanoma adhesion molecule) | 10.791 |
| NM_002545 | OPCML (opioid binding protein/cell adhesion molecule-like) | 10.561 |
| NM_002933 | RNASE1 (ribonuclease, RNase A family, 1) | 6.81 |
| NM_001993 | F3 (coagulation factor III) | 6.29 |
| NM_005433 | YES1(v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | 5.9161 |
| NM_016525 | UBAP (ubiquitin associated protein) | 5.876 |
| NM_001428 | ENO1 (enolase 1) | 5.6921 |
| S68616 | SLC9A1 (Na+/H+ exchanger NHE-1 isoform) | 5.484 |
| NM_003254 | TIMP1 (tissue inhibitor of metalloproteinase 1) | 5.291 |
| NM_005903 | MADH5 (mothers against decapentaplegic, homolog 5) | 5.083 |
| U82828 | ATM (ataxia telangiectasia) | 0.2051 |
| NM_006343 | MERTK (c-mer proto-oncogene tyrosine kinase) | 0.19 |
| NM_002985 | SCYA5 (small inducible cytokineA5) | 0.191 |
| NM_005348 | HSPCA (heat shock 90kD protein 1, alpha) | 0.164 |
| NM_003968 | UBE1C (ubiquitin-activating enzyme E1C) | 0.161 |
| NM_004374 | COX6C (cytochrome c oxidase subunit VIc) | 0.131 |
| NM_002990 | SCYA22 (small inducible cytokine subfamily A, member22) | 0.129 |
| NM_005781 | ACK1 (activated p21cdc42Hs kinase) | 0.1281 |
| NM_005139 | ANXA3 (annexin A3) | 0.124 |
| AF053630 | SERPINB1(serine proteinase inhibitor, clade B, member 1) | 0.123 |
| NM_012090 | MACF1 (microtubule-actin crosslinking factor 1) | 0.098 |
| XM_042551 | CAMK2A (calcium/calmodulin-dependent protein kinase) | 0.0645 |
| NM_000909 | NPY1R (neuropeptide Y receptor Y1) | 0.02521 |
| NM_015230 | CENTD1 (centaurin, delta 1) | 0.024 |
| NM_004958 | FRAP1 (FK506 binding protein 12-rapamycin associated protein 1) | 0.0205 |
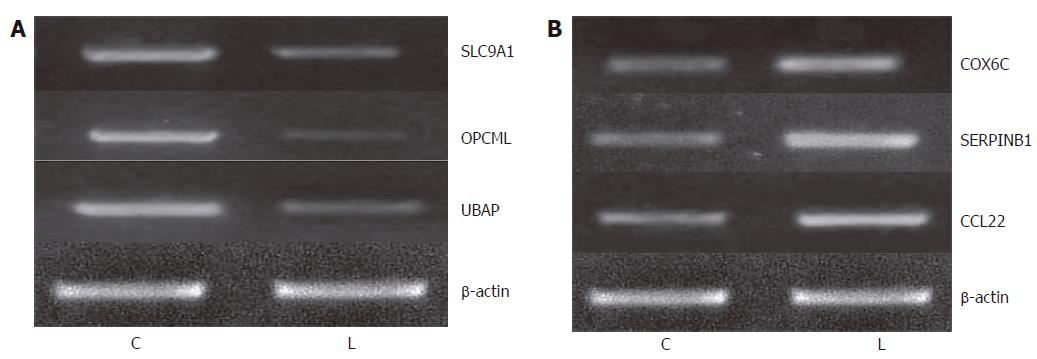
To examine the reliability of microarray data, we confirmed our data at the level of mRNA and protein. First, we selected 3 up-regulated genes (SLC9A1, OPCML and UBAP) and 3 down-regulated genes (COX6C, SERPINB1 and CCL22) to measure their expression levels by semiquantitative RT-PCR. To obtain truly comparable results, we used the unamplified total RNA (from the same batch used for array hybridizations) as the template. The results were very similar to the microarray data on these genes.
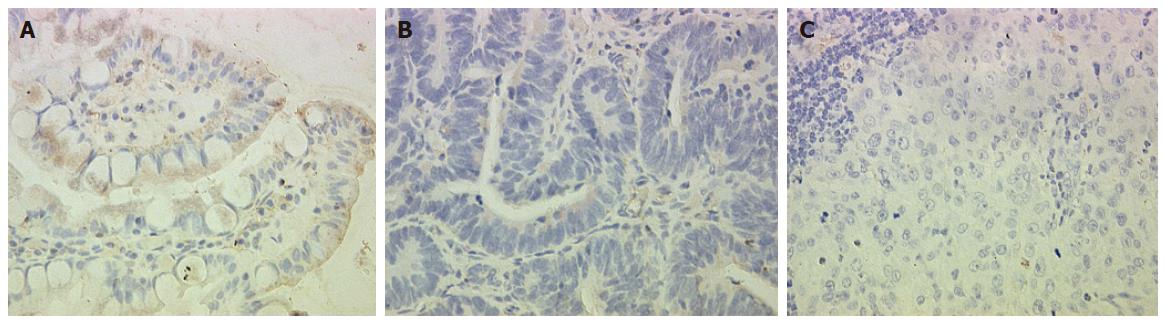
To confirm our data at the protein level, we performed immunohistochemical analysis of OPCML. The results were in parallel with the differential expression pattern detected by cDNA microarray and RT-PCR (Figure 2).
Overall, the above results demonstrated that the samples obtained by P27-based amplification well reflected the status of the original RNA in a proportional manner, and supported the reliability of our expression data (Figure 3).
By semiquantitative RT-PCR, we further evaluated the expression levels of 25 selected genes, including 10 up-regulated and 15 down-regulated genes, in microdissected normal gastric epithelial cells from 30 healthy individuals, primary gastric cancer cells from 36 patients and lymph node metastatic cells from the remaining 22 patients. First, we investigated the expression levels of these genes in primary gastric cancer and lymph node metastatic cells. The results showed that the expression of 12 genes had the same pattern in > 50% (12/22) patients as revealed by cDNA microarray.
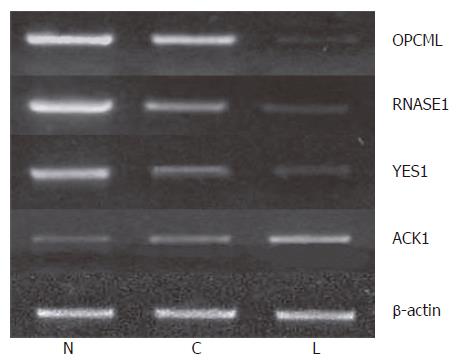
We further measured the expression levels of these 12 genes in paired normal gastric epithelium and primary carcinoma samples from the same 22 patients. We found that the expression pattern of 3 genes (OPCML, RNASE1 and YES1) was similar to that of tumor suppressor genes in > 50% (8/15) patients. For example, the expression level of these genes was the highest in normal gastric epithelium, which was decreased in primary carcinoma and further decreased compared to that in primary gastric cancer patients with lymph node metastasis and normal controls (P < 0.05). Meanwhile, the expression level of ACK1 demonstrated the opposite tendency, the pattern of which was similar to that of oncogene in > 50% (12/22) patients (Figure 4). The expression pattern of OPCML was also confirmed at the protein level by immunohistochemical staining (Figure 2).
cDNA microarrays allow an effective investigation of functional genomics. However, the existence of bulky surrounding cells produces much useless noise information because of its high sensitivity[9,10]. Therefore, selection of cancer cells using LMD is of indispensable value in combination with the cDNA microarray. The LMD system used in this study integrates a UV laser of 337 nm wavelength with an upright microscope. The ultraviolet laser microbeam causes dissection by local photolysis of the supporter membrane and tissue section due to the high photon density of the microbeam rather than by local heating or coagulation. The cut sample falls down into PCR tubes placed underneath by gravity without any mechanical contact or further destroying energy and the integrity of the extracted mRNA is maximally kept.
Metastasis of cancer is a highly selective sequential step which favors the survival of a subpopulation of metastatic cells preexisting within the primary tumor mass to produce clinically relevant metastases. The metastatic cells exhibit a complex phenotype that is regulated by transient or permanent changes in different genes at the DNA and/or mRNA level(s). This has also been proved in gastric cancer by other researchers[8,9,11].
Differential expression profiles of laser microdissected primary gastric cancer cells and lymph node metastatic cells were discovered using cDNA microarray consisting of 13824 genes, demonstrating that analysis of gene-expression profiles can be performed using LMD, P27-based RNA amplification and cDNA microarray[1,11]. Although the majority of these genes have been implicated in various aspects of tumor biology, few are associated with gastric cancer.
Among the above genes, some may be differentially expressed because of different living environments[5,12-17]. Therefore, to further confirm and screen the results of cDNA microarray, we measured the expression levels of 25 selected genes in 22 patients by semiquantitative RT-PCR. These target cells were collected by LMD, and the normal gastric epithelium was included. As a result, we identified 4 genes, the expression level of which was different not only between primary carcinoma and metastatic lymph nodes (the same results as cDNA microarray), but also between normal gastric epithelium and primary tumor, suggesting that these four genes play a key role in the tumorigenesis and metastasis of gastric cancer. The expression pattern of 3 genes (OPCML, RNASE1 and YES1) is similar to that of tumor suppressor genes. For example, the expression of these genes is the highest in normal gastric epithelium, which is decreased in primary carcinoma, and further decreased in metastatic lymph nodes. OPCML encodes a member of the IgLON subfamily in the immunoglobulin protein superfamily and acts as a GPI-anchored protein. It was reported that OPCML has tumor-suppressor function in epithelial ovarian cancer[11,18-20], which is in accordance with our findings in gastric cancer. Interestingly, YES1 is the cellular homolog of a virus oncogene associated with esophageal tumorigenesis[12,21-24], which is contrary to our results. The RNAS gene encodes a member of the pancreatic-type of secretory ribonuclease, a subset of the ribonuclease A superfamily, and has no relationship with human cancers.
In this study, the expression pattern of gene ACK1 was different from that of the above mentioned genes, but similar to that of oncogenes. ACK1 encodes a tyrosine kinase that binds to Cdc42Hs in its GTP-bound form and inhibits intrinsic and GTPase-activating protein (GAP)-stimulated GTPase activity of Cdc42Hs. It is directly linked to a tyrosine phosphorylation signal transduction pathway, but its effect on tumor progression has not been reported[12-17].
Both tumor suppressor gene and oncogene are important target molecules in clinical diagnosis and treatment of malignant gastric tumors. The different expressions of these four genes have not been reported during the progression of gastric cancer.
In conclusion, analysis of gene expression profiles by LMD, P27-based amplification, and cDNA microarray can provide useful information for clarifying the mechanism underlying the development and metastasis of gastric cancer[25-29], not only revealing the differentially expressed genes in progression of gastric cancer, but also providing information for identifying novel diagnostic and therapeutic targets.
S- Editor Wang J L- Editor Wang XL E- Editor Bi L
| 1. | Zhu JS, Shen B, Chen JL, Chen GQ, Yu XH, Yu HF, Zhu ZM. Molecule action mechanisms of NM-3 on human gastric cancer SGC-7901 cells in vivo or in vitro. World J Gastroenterol. 2003;9:2366-2369. [PubMed] |
| 2. | Yasui W, Oue N, Ono S, Mitani Y, Ito R, Nakayama H. Histone acetylation and gastrointestinal carcinogenesis. Ann N Y Acad Sci. 2003;983:220-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 3. | Wang ZN, Xu HM, Jiang L, Zhou X, Lu C, Zhang X. Expression of survivin mRNA in peritoneal lavage fluid from patients with gastric carcinoma. Chin Med J (Engl). 2004;117:1210-1217. [PubMed] |
| 4. | Portera CA Jr, Berman RS, Ellis LM. Molecular determinants of colon cancer metastasis. Surg Oncol. 1998;7:183-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 295] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 6. | Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1818] [Cited by in RCA: 1696] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 7. | Böhm M, Wieland I, Schütze K, Rübben H. Microbeam MOMeNT: non-contact laser microdissection of membrane-mounted native tissue. Am J Pathol. 1997;151:63-67. [PubMed] |
| 8. | Liotta L, Petricoin E. Molecular profiling of human cancer. Nat Rev Genet. 2000;1:48-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 272] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 9. | Mori M, Mimori K, Yoshikawa Y, Shibuta K, Utsunomiya T, Sadanaga N, Tanaka F, Matsuyama A, Inoue H, Sugimachi K. Analysis of the gene-expression profile regarding the progression of human gastric carcinoma. Surgery. 2002;131:S39-S47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Zhu JS, Wang L, Cheng GQ, Li Q, Zhu ZM, Zhu L. Apoptosis mechanisms of human gastric cancer cell line MKN-45 infected with human mutant p27. World J Gastroenterol. 2005;11:7536-7540. [PubMed] |
| 11. | Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34:337-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 139] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Nakakuki K, Imoto I, Pimkhaokham A, Fukuda Y, Shimada Y, Imamura M, Amagasa T, Inazawa J. Novel targets for the 18p11.3 amplification frequently observed in esophageal squamous cell carcinomas. Carcinogenesis. 2002;23:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Nan KJ, Jing Z, Gong L. Expression and altered subcellular localization of the cyclin-dependent kinase inhibitor p27Kip1 in hepatocellular carcinoma. World J Gastroenterol. 2004;10:1425-1430. [PubMed] |
| 14. | Maeshima Y, Manfredi M, Reimer C, Holthaus KA, Hopfer H, Chandamuri BR, Kharbanda S, Kalluri R. Identification of the anti-angiogenic site within vascular basement membrane-derived tumstatin. J Biol Chem. 2001;276:15240-15248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 163] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Bryja V, Pacherník J, Faldíková L, Krejcí P, Pogue R, Nevrivá I, Dvorák P, Hampl A. The role of p27(Kip1) in maintaining the levels of D-type cyclins in vivo. Biochim Biophys Acta. 2004;1691:105-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1411] [Cited by in RCA: 1442] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 17. | Liu E, Li X, Yan F, Zhao Q, Wu X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J Biol Chem. 2004;279:17283-17288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 133] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Guo W, Shang F, Liu Q, Urim L, West-Mays J, Taylor A. Differential regulation of components of the ubiquitin-proteasome pathway during lens cell differentiation. Invest Ophthalmol Vis Sci. 2004;45:1194-1201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 19. | Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 389] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 20. | Kudo Y, Kitajima S, Sato S, Ogawa I, Miyauchi M, Takata T. Transfection of p27(Kip1) threonine residue 187 mutant type gene, which is not influenced by ubiquitin-mediated degradation, induces cell cycle arrest in oral squamous cell carcinoma cells. Oncology. 2002;63:398-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 21. | Hurteau JA, Brutkiewicz SA, Wang Q, Allison BM, Goebl MG, Harrington MA. Overexpression of a stabilized mutant form of the cyclin-dependent kinase inhibitor p27(Kip1) inhibits cell growth. Gynecol Oncol. 2002;86:19-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 22. | Koguchi K, Nakatsuji Y, Nakayama K, Sakoda S. Modulation of astrocyte proliferation by cyclin-dependent kinase inhibitor p27(Kip1). Glia. 2002;37:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Zhang D, Holmes WF, Wu S, Soprano DR, Soprano KJ. Retinoids and ovarian cancer. J Cell Physiol. 2000;185:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 24. | Koh TY, Park SW, Park KH, Lee SG, Seol JG, Lee DW, Lee CT, Heo DS, Kim KH, Sung MW. Inhibitory effect of p27KIP1 gene transfer on head and neck squamous cell carcinoma cell lines. Head Neck. 2003;25:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Sasaki T, Katayose Y, Suzuki M, Yamamoto K, Shiraso S, Mizuma M, Unno M, Takeuchi H, Lee CT, Matsuno S. Adenovirus expressing mutant p27kip1 enhanced apoptosis against cholangiocarcinoma than adenovirus-p27kip1 wild type. Hepatogastroenterology. 2004;51:68-75. [PubMed] |
| 26. | Park KH, Seol JY, Kim TY, Yoo CG, Kim YW, Han SK, Shim YS, Lee CT. An adenovirus expressing mutant p27 showed more potent antitumor effects than adenovirus-p27 wild type. Cancer Res. 2001;61:6163-6169. [PubMed] |
| 27. | Park KH, Lee J, Yoo CG, Kim YW, Han SK, Shim YS, Kim SK, Wang KC, Cho BK, Lee CT. Application of p27 gene therapy for human malignant glioma potentiated by using mutant p27. J Neurosurg. 2004;101:505-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 28. | Winteringham LN, Kobelke S, Williams JH, Ingley E, Klinken SP. Myeloid Leukemia Factor 1 inhibits erythropoietin-induced differentiation, cell cycle exit and p27Kip1 accumulation. Oncogene. 2004;23:5105-5109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Takeda A, Osaki M, Adachi K, Honjo S, Ito H. Role of the phosphatidylinositol 3'-kinase-Akt signal pathway in the proliferation of human pancreatic ductal carcinoma cell lines. Pancreas. 2004;28:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |