Published online Sep 7, 2006. doi: 10.3748/wjg.v12.i33.5281
Revised: April 28, 2006
Accepted: May 22, 2006
Published online: September 7, 2006
AIM: To shed light on the possible role of mismatch repair gene Mlh3 in familial esophageal cancer (FEC).
METHODS: A total of 66 members from 10 families suggestive of a genetic predisposition to hereditary esophageal cancer were screened for germline mutations in Mlh3 with denaturing high performance liquid chromatography (DHPLC), a newly developed method of comparative sequencing based on heteroduplex detection. For all samples exhibiting abnormal DHPLC profiles, sequence changes were evaluated by cycle sequencing. For any mutation in family members, we conducted a segregation study to compare its prevalence in sporadic esophageal cancer patients and normal controls.
RESULTS: Exons of Mlh3 in all samples were successfully examined. Overall, 4 missense mutations and 3 polymorphisms were identified in 4 families. Mlh3 missense mutations in families 9 and 10 might be pathogenic, but had a reduced penetrance. While in families 1 and 7, there was no sufficient evidence supporting the monogenic explanations of esophageal cancers in families. The mutations were found in 33% of high-risk families and 50% of low-risk families.
CONCLUSION: Mlh3 is a high risk gene with a reduced penetrance in some families. However, it acts as a low risk gene for esophageal cancer in most families. Mutations of Mlh3 may work together with other genes in an accumulated manner and result in an increased risk of esophageal tumor. DHPLC is a robust and sensitive technique for screening gene mutations.
- Citation: Liu HX, Li Y, Jiang XD, Yin HN, Zhang L, Wang Y, Yang J. Mutation screening of mismatch repair gene Mlh3 in familial esophageal cancer. World J Gastroenterol 2006; 12(33): 5281-5286
- URL: https://www.wjgnet.com/1007-9327/full/v12/i33/5281.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i33.5281
Esophageal carcinoma (EC) is one of the most common malignant tumors in China[1,2]. Although various therapeutic strategies have been improved in recent years, the 5-year survival remains poor, therefore, early diagnosis and treatment of EC are still urgently needed[3,4].
Current studies showed that multiple factors and steps contribute to its tumorigenesis, however the definite mechanisms remain to be determined yet[5,6]. While in cancer families, genetic predisposition may play a more important role in carcinogenesis than other factors[7]. Genetic susceptibility can be identified by detecting germ-line mutation of certain genes from peripheral blood.
Denaturing high performance liquid chromatography (DHPLC) is a newly developed technique suitable for the detection of heteroduplex mutations. DHPLC has the advantages of being highly automated, sensitive, and could be used for screening of gene mutations on a large scale[8].
Great attention has been paid to mismatch repair (MMR) genes due to their association with hereditary non-polyposis colorectal cancer (HNPCC)[9]. Mlh3 gene was first identified in 2000, which is a putative MMR gene normally expressed in multiple epithelia. It is located on 14q24.3 with a coding length of 4.3 kb, and is composed of 12 exons, of which exon 1 is 3.3 kb, accounting for 75%[10]. Some investigations showed that it might play a certain role in the tumorigenesis of colorectal cancer[11-14]. However, some studies indicate that there is no strong association between currently known Mlh3 variants and colorectal cancer predisposition risk[15,16]. A recent study on familial gastric cancer showed that Mlh3 can act as a low risk gene[17].
Therefore, in an attempt to further evaluate its possible role in the cancer family, we performed mutation screening of Mlh3 in familial esophageal cancer (FEC) with DHPLC.
A total of 10 families with a hereditary background of esophageal carcinoma were collected (Table 1). The criteria were set as: at least 2 generations with esophageal cancers, at least two affected members, first-degree relatives, and carcinogenesis at earlier ages (< 50 years old). In addition to 66 members from these 10 esophageal families, 96 cases of sporadic esophageal cancer and 96 normal controls were recruited from the Department of Thoracic Surgery, First Hospital, China Medical University. Blood samples were collected from the members of each family after giving their informed consent.
| Type | Family |
| TCR1 | 1, 5, 8, 10 |
| MCR2 | 2, 3 ,4, 6, 7, 9 |
DNA was extracted using the standard phenol/chloroform extraction protocols.
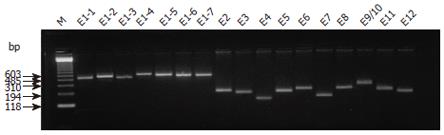
Seventeen primers were used for a total of twelve exons in Mlh3. Exon 1 was divided into seven overlapping fragments, and exons 8 and 9 were amplified together because of their small size and the small intron between them. The length of PCR fragments, primer sequences and corresponding annealing temperature for each fragment are shown in Table 2. All products were examined for specificity and quantity by 2% agarose gel electrophoresis.
| Primer sequence | Length | Annealing temperature |
| (bp) | (°C) | |
| MLH3 F1-1: TCC AGT CAG AGA AGG AAA CCA | 524 | Touch down |
| MLH3 R1-1: ACA GGA AGC TGG TAA AAT AG | 58-51 | |
| MLH3 F1-2: CTG ATG TGA CTA GAG CAA GCG | 552 | Touch down |
| MLH3 R1-2: CAT CAT ACT CAC AGA ATT GGC AC | 58-51 | |
| MLH3 F1-3: ATT CAA GTC TTC GGC ACC | 512 | Touch down |
| MLH3 R1-3: TTT GTT TTG TAA AGA TGG CTC TG | 58-51 | |
| MLH3 F1-4: GGG ATT CAG AAG CTA CCA | 602 | Touch down |
| MLH3 R1-4: TGA ATG TTC TGT TTC AGT TGA TTT | 58-51 | |
| MLH3 F1-5: GGG CGA GTT AAA TTA TGT TCC A | 562 | Touch down |
| MLH3 R1-5: CTT GAA GAC TGA GAT TGG TAG TGA | 58-51 | |
| MLH3 F1-6: TGG GAA GGT TGA AAA TCC TC | 557 | Touch down |
| MLH3 R1-6:AGG AAT TAT CCT GTG TGG CAG | 58-51 | |
| MLH3 F1-7: CAG AGA ATG GTG TCA TCC CAA | 551 | Touch down |
| MLH3 R1-7: CCT TGT CCA GCA TTC CCA T | 58-51 | |
| MLH3 F2: TGT CTT GAC TCA GTT TGT GCA G | 247 | 62 |
| MLH3 R2: ACG ATG TGT ACT GTG TGC CC | ||
| MLH3 F3: TGG TTC TGG ATG CCA ACT TT | 229 | 59 |
| MLH3 R3: ATT TCA GTC TGG GCA ACA GG | ||
| MLH3 F4: CAA TTA TAT TTT GCT GAG TC | 158 | 52 |
| MLH3 R4: ATG AGA TTT TGA AGT TAA TC | ||
| MLH3 F5: CCC AGT CTC AAA GAA AGG AGT | 239 | 57 |
| MLH3 R5: AGC TGG TTA GTC ATT CAG GC | ||
| MLH3 F6: CAT GAT GGT TGT CGT CTT GC | 185 | 60 |
| MLH3 R6: GGT GTA CTG ATT CTG CTG GGA | ||
| MLH3 F7: TTC CCT TCC TAC TCT TAA CCC A | 265 | 59 |
| MLH3 R7: TGT AAC CTC TCT TGG TCT CAT CTG | ||
| MLH3 F8: TTT GGA ACC AGT AGT GAA GTG C | 267 | 57 |
| MLH3 R8: CAG CAA TTT CCT TAA CAT CTG C | ||
| MLH3 F9/10: CGT AGA TTA AAG CCG ATT TTC | 329 59 | |
| MLH3 R9/10:TGT ACC CTC TGC CTC TTT CG | ||
| MLH3 F11: GTC AGC ATT GGT TTC CCA CT | 251 | 59 |
| MLH3 R11: AAA CTT TGC TCC CTC CTG CT | ||
| MLH3 F12: GCC CAG CCT GTA TGC TAC CT | 226 | 59 |
| MLH3 R12: CAG TGA CAC TCC CTT TGT TCC |
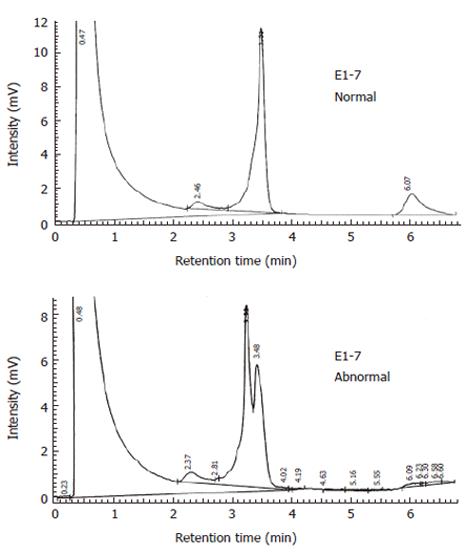
PCR products were denatured at 95°C, and gradually cooled down to room temperature to enable efficient formation of heteroduplexes. DHPLC was carried out in a transgenomic wave DNA fragment analysis system, an automated DHPLC instrumentation equipped with a DNASep column. Abnormal elution profiles were identified by visual inspection of the chromatogram on the basis of the appearance of one or more additional earlier eluting peaks. Temperatures used in DHPLC analysis have been described elsewhere[14].
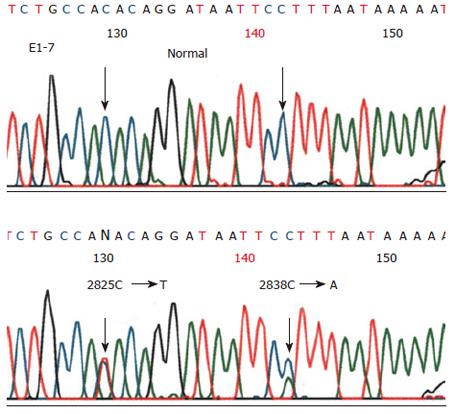
Genomic DNA was re-amplified with DHPLC primers. After purification, DNA sequence changes were evaluated by direct sequencing in both directions. After treatment with ethanol, cycle sequencing products were analyzed on an ABI PRISM® 377 DNA sequencer.
For any specific variant of Mlh3 gene, we also detected its percentage in 96 cases of sporadic esophageal cancer and 96 normal controls in addition to the segregation study in families. If there was no such mutation in sporadic cancer or in normal controls, then this mutation might be pathogenic in this family. On the other hand, it might be a polymorphism not associated with the disease.
All the exons were successfully amplified (Figure 1), analyzed with DHPLC (Figure 2) and sequenced (Figure 3). Four missense mutations and 3 polymorphisms were identified in 4 families (Tables 3 and 4). One variant C2531T (Pro8441Leu) had almost the same prevalence in familial cancer, sporadic cancer and controls. One silent variant C2838A (Ser947Ser) and an intronic variant (between exons 9 and 10) A→G, were considered polymorphisms (Table 4). All variants were not in the conserved homologus regions of the MutL protein at the NH2 and COOH terminals, and none of the variants was evolutionarily conserved in yeast or E. coli[18,19].
| Exon | Nucleotide | Amino | Family | Family | Cosegregation | Frequency in | Frequency in sporadic | Frequency in normal |
| change1 | change | type | FEC n (%) | EC n (%) | control n (%) | |||
| 1 | A2173C | Asn725His | 9 | MCR | Yes | 4/66 (6.1) | 0/94 (0) | 0/96 (0) |
| 1 | C2825T | Thr9421Ile | 10 | TCR | Yes | 3/66 (4.5) | 1/95 (1.0) | 0/96 (0) |
| 7 | T3826C | Trp1276Arg | 1 | TCR | No | 3/66 (4.5) | 0/96 (0) | 0/96 (0) |
| 12 | G4335A | Gln1445Glu | 7 | MCR | No | 6/66 (9.1) | 0/95 (0) | 0/96 (0) |
| Exon | Nucleotide change1 | Amino change | Family |
| 1 | C2531T | Pro8441Leu | Many |
| 1 | C2838A | Ser947Ser | Many |
| 9/10 | IVS9 + 66A→G | Unknown | Many |
The families with identified mutations were of two types, high risk families consisting of more than three affected first degree relatives (MCR), and low risk families consisting of two affected first degree relatives (TCR) (Table 1). There were 6 high risk families and 4 low risk families. Two missense mutations (T3826C and C2825T) were found in two low risk families (families 1 and 10), the other two variants (G4335A and A2173C) were observed in high risk families (families 7 and 9). Mutations were found in 33% (2/6) of the high risk families and 50% (2/4) of the low risk families.
To elucidate the pathogenic nature of these 4 variants, we also detected their prevalence in sporadic cases and normal controls (Table 3). The data demonstrated that the frequency in families was far higher than that in sporadic cases and normal controls, suggesting that they might contribute to the disease in these families.
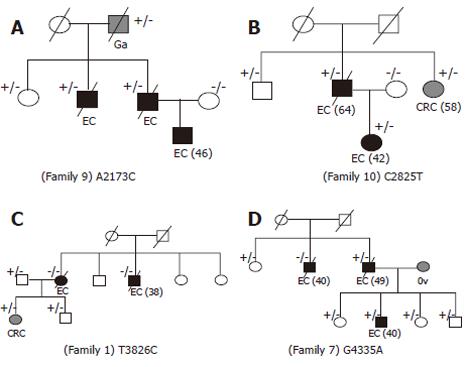
Missense mutation A2173C was found in 3 patients with esophageal cancer, one gastric caner patient and one 76-year old unaffected relative in family 9 (Figure 4A), suggesting that the mutation could be inherited from the grandfather. Furthermore, no such variant was detected in controls and sporadic cases, suggesting that the mutation could be associated with the occurrence of esophageal cancer, but might have a reduced penetrance.
Missense mutation C2825T was found in family 10 (Figure 4B). The mutation was shared by the father and daughter with esophageal cancer, unaffected brother (66 years old) and sister with colon cancer, which was consistent with the mutation associated with the disease but having a reduced penetrance. Moreover, the mutation was found in 4.5% of family members and 1.0% of sporadic cases, but not in controls.
Variant T3826C(Trp1276Arg)in exon 7 was found in family 1 (Figure 4C). However, the mutation did not segregate well with the disease. Two affected members with esophageal cancer had no such mutation. While one female with colon cancer and two males with no colon cancer had this mutation. T3826C was not detected in controls and sporadic cases.
Variant G4335A was found in a male patient with esophageal cancer and his affected son in family 7, but not in his brother with esophageal cancer, whereas his unaffected sisters and three healthy children shared this mutation (Figure 4D). G4335A was not detected in sporadic cases and normal controls.
Among the esophageal cancer families, Mlh3 mutation A2173C in family 9 and C2825T in family 10 could be causative, but had a reduced penetrance. These mutations were segregated with the disease, and neither of them was found in sporadic cases and normal controls. In variant T3826C in family 1 and G4335A in family 7, there was a lack of evidence of the monogenic high risk predispo-sition to esophageal cancer associated with these identified variants, because there was no clear association between mutation and the disease. However, these variants were more prevalent in families but not in sporadic cases and the controls, indicating a possible pathogenic nature in these families. Surprisingly, mutations were found in 50% (2/4) of low risk families and in 33% of high risk families (2/6).
The mismatch repair system is composed of a highly diverse group of proteins that interact with numerous DNA structures during DNA repair and replication[20].
The MutL homologue, Mlh3 gene was first identified as a new member of the DNA MMR gene in yeast, and has been mapped to the region of the mouse complex trait locus, colon cancer susceptibility I[10]. It is highly conserved in evolution. The protein encoded by its carboxyl-terminal may interact with the currently known mismatch repair gene MLH1, and is very similar to Mlh3p in yeast, indicating that it may play a role in the DNA mismatch repair system[10]. It is essential not only for DNA repair and microsatellite stability, but also for meiosis. In S. cerevisiae, Mlh1-Mlh3 complex acts competitively in a distinct way to promote crossing over during meiosis along with the Msh4-Msh5 and Mus81-Mms4 complexes[21]. The significant heterogeneity in localization of the MutL homologues, Mlh1 and Mlh3, directly results in defective crossing over during meiotic recombination in prophase I, and consequently 30% of human oocytes are predisposed to aneuploidy[22].
Furthermore, Wu et al[23] used knockout mlh3 (-/-) mice to address the role of Mlh3 in class switch DNA recombination (CSR) and somatic hypermutation (SHM), and found that Mlh3 deficiency alters both CSR and SHM, suggesting that the MMR Mlh3 protein plays a role in both CSR and SHM.
A recent study reported that Mlh3 contributes to tumor suppression in mice. Mlh3 deficiency causes microsatellite instability, impaired DNA-damage response, and increased gastrointestinal tumor susceptibility[24]. There is evidence that mismatch repair gene mutations is significantly associated with an increased risk of developing colorectal cancer[25] and that the mismatch repair gene Mlh3 plays an important role in DNA repair.
There is evidence that mismatch repair gene Mlh3 plays an increasingly important role in DNA repair after replication, microsatellite stability, meiosis and tumor suppression.
In this study, among the 66 members in 10 esophageal cancer families, 4 missense mutations and 3 possible polymorphisms were idenbtified. Even though all the identified mutations were not in the conserved region and none of them changed a codon conserved in yeast and E. coli, their possible role in tumorigenesis cannot be ruled out. In fact, many reported missense mutations are not within the conserved region of hMLH1, another important mismatch repair gene[26].
Current investigations have been mainly focused on its susceptibility to colorectal cancer (CRC)[11-16]. By using DHPLC to screen Mlh3 mutations in 70 families with likely genetic predisposition to colon cancer, Liu et al[14] found that the frequency of its germ-line variants , including one frameshift mutation, 10 missense mutations and 5 polymorphisms was high (23%). By analyzing 30 CRC cases for germline mutations by sequencing, Hienonen et al[15] have found 5 missense variants, 4 of which were also found in cancer-free controls. The only remaining variant does not appear to be an attractive candidate for a disease-associated mutation because the amino acid change is located outside the conserved residues. Furthermore, they have not found the reported frameshift mutation in the 30 CRC cases or in 700 cancer-free controls. While it is a difficult task to exclude the role of Mlh3 in HNPCC, their study could not confirm the role of Mlh3 in CRC predisposition. Similarly, De Jong et al[16] have found two variants (S845G and P844L) in Dutch patients with suspected HNPCC, but further investigation has failed to demonstrate an association between the two variants and colorectal cancer risk. Zhao et al[17] performed an investigation on Mlh3 in 16 suggestive hereditary gastric cancer families with DHPLC and indentified 5 missence mutations. However, no strong association has been verified between these 5 variants and gastric cancer risk, indicating that Mlh3 probably acts as a low risk gene in familial gastric cancer.
To establish whether the mutations segregate with disease in a family, we tried to collect DNA samples from members of each family. Two variants (A2173C and C2825T) showed a segregation pattern consistent with a monogenetic risk factor in families 9 and 10. Additionally, neither of them was detected among sporadic esophageal cancer cases and normal controls. However, because the sizes of these families were small, and the affected individuals were all first-degree relatives, Mlh3 could not be defined as a high penetrant predisposing FEC gene. The other variants (T3826C and G4335A) had no clear segregation pattern consistent with the disease, even the frequencies in families were higher than those in sporadic cases and controls.
Although the mutations of Mlh3 identified in our study did not provide sufficient evidence supporting a monogenetic explanation for the familial aggregation of EC, we would still like to propose that some or all of these mutations might work as low risk genes, perhaps in an accumulated manner, resulting in increased risk for EC. We believe that the majority of EC are caused by some low risk genes, which act on their own or interact in an additive manner. Therefore, the monogenetic susceptibility is still suggested for the disease in high risk families (family 7 and 9), while multiple genetic inheritance of susceptibility could be the reason in low risk families (family 1 and 10). Chen et al[24] found mismatch repair Mlh3 and Pms2 double-deficient mice have tumor susceptibility, shorter life span, microsatellite instability, and DNA-damage response phenotypes. Al-Tassan[27] found that in MYH, another DNA repair gene, several missense mutations act in a recessive manner and cause colorectal cancers in three sibs, with compound heterozygous missense mutations in this gene. Some mutations were also found as heterozygous missense mutations in healthy members of the same family and normal controls. Although, the predisposition seems to be inherited in a recessive manner, it is possible that heterozygous mutations in this gene also act in an additive manner as a low risk gene. And in our previous study on colorectal cancer families, we found a hMLH3 mutation segregated with disease together with a missense mutation in hMSH2[14] .The missense mutations of both Mlh3 and Msh2 contributes to the failure of mismatch repair-mediated tumor suppression, resulting in the family aggregation of colorectal cancer.
Additionally, DHPLC analysis is very sensitive, robust and reproducible. Nevertheless, the relatively high expensive apparatus and related reagents have limited its wide use.
Based on previous reports and our results, the Mlh3 gene might occasionally appear as a high risk gene predisposing to EC, while in most cases it works as a low risk mismatch repair gene, contributing to the increased risk of developing EC. Despite our findings, much is yet to be learned about the molecular basis of correlations between genetic changes and clinical features of the disease. DHPLC is a sensitive and robust technique for screening gene mutations.
S- Editor Wang J L- Editor Wang XL E- Editor Ma WH
| 1. | Su M, Lu SM, Tian DP, Zhao H, Li XY, Li DR, Zheng ZC. Relationship between ABO blood groups and carcinoma of esophagus and cardia in Chaoshan inhabitants of China. World J Gastroenterol. 2001;7:657-661. [PubMed] |
| 2. | Gu ZD, Chen KN, Li M, Gu J, Li JY. Clinical significance of matrix metalloproteinase-9 expression in esophageal squamous cell carcinoma. World J Gastroenterol. 2005;11:871-874. [PubMed] |
| 3. | An JY, Fan ZM, Zhuang ZH, Qin YR, Gao SS, Li JL, Wang LD. Proteomic analysis of blood level of proteins before and after operation in patients with esophageal squamous cell carcinoma at high-incidence area in Henan Province. World J Gastroenterol. 2004;10:3365-3368. [PubMed] |
| 4. | Lin YC, Wu MY, Li DR, Wu XY, Zheng RM. Prognostic and clinicopathological features of E-cadherin, alpha-catenin, beta-catenin, gamma-catenin and cyclin D1 expression in human esophageal squamous cell carcinoma. World J Gastroenterol. 2004;10:3235-3239. [PubMed] |
| 5. | Li J, Nu ZX, Wang LD. The Molecular Biology of esophageal carcinoma. Shijie Huaren Xiaohua Zazhi. 1997;5:189-191. |
| 6. | Wang DX, Li W. Advances in esophageal neoplasms etiology. Shijie Huaren Xiaohua Zazhi. 2000;8:1029-1030. |
| 7. | Abbas A, Delvinquiere K, Lechevrel M, Lebailly P, Gauduchon P, Launoy G, Sichel F. GSTM1, GSTT1, GSTP1 and CYP1A1 genetic polymorphisms and susceptibility to esophageal cancer in a French population: different pattern of squamous cell carcinoma and adenocarcinoma. World J Gastroenterol. 2004;10:3389-3393. [PubMed] |
| 8. | Arnold N, Gross E, Schwarz-Boeger U, Pfisterer J, Jonat W, Kiechle M. A highly sensitive, fast, and economical technique for mutation analysis in hereditary breast and ovarian cancers. Hum Mutat. 1999;14:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 9. | Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801-818. [PubMed] |
| 10. | Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, Elliott RM, Collins FS. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 217] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 11. | Lipkin SM, Wang V, Stoler DL, Anderson GR, Kirsch I, Hadley D, Lynch HT, Collins FS. Germline and somatic mutation analyses in the DNA mismatch repair gene MLH3: Evidence for somatic mutation in colorectal cancers. Hum Mutat. 2001;17:389-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Loukola A, Vilkki S, Singh J, Launonen V, Aaltonen LA. Germline and somatic mutation analysis of MLH3 in MSI-positive colorectal cancer. Am J Pathol. 2000;157:347-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Wu Y, Berends MJ, Sijmons RH, Mensink RG, Verlind E, Kooi KA, van der Sluis T, Kempinga C, van dDer Zee AG, Hollema H. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet. 2001;29:137-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 106] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 14. | Liu HX, Zhou XL, Liu T, Werelius B, Lindmark G, Dahl N, Lindblom A. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63:1894-1899. [PubMed] |
| 15. | Hienonen T, Laiho P, Salovaara R, Mecklin JP, Järvinen H, Sistonen P, Peltomäki P, Lehtonen R, Nupponen NN, Launonen V. Little evidence for involvement of MLH3 in colorectal cancer predisposition. Int J Cancer. 2003;106:292-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | de Jong MM, Hofstra RM, Kooi KA, Westra JL, Berends MJ, Wu Y, Hollema H, van der Sluis T, van der Graaf WT, de Vries EG. No association between two MLH3 variants (S845G and P844L)and colorectal cancer risk. Cancer Genet Cytogenet. 2004;152:70-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 17. | Zhao CH, Liu HX, Bu XM. Mutation of mismatch repair gene hMLH3 in familial gastric cancer. Shijie Huaren Xiaohua Zazhi. 2004;12:1030-1033. |
| 18. | Kondo E, Horii A, Fukushige S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 2001;29:1695-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 298] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 20. | Hoffmann ER, Borts RH. Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res. 2004;107:232-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 21. | Argueso JL, Wanat J, Gemici Z, Alani E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics. 2004;168:1805-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 22. | Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am J Hum Genet. 2005;76:112-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 23. | Wu X, Tsai CY, Patam MB, Zan H, Chen JP, Lipkin SM, Casali P. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J Immunol. 2006;176:5426-5437. [PubMed] |
| 24. | Chen PC, Dudley S, Hagen W, Dizon D, Paxton L, Reichow D, Yoon SR, Yang K, Arnheim N, Liskay RM. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662-8670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 25. | Mitchell RJ, Farrington SM, Dunlop MG, Campbell H. Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am J Epidemiol. 2002;156:885-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 99] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1448] [Cited by in RCA: 1359] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 27. | Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR. Inherited variants of MYH associated with somatic G: C--> T: A mutations in colorectal tumors. Nat Genet. 2002;30:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 960] [Cited by in RCA: 930] [Article Influence: 40.4] [Reference Citation Analysis (0)] |