Published online Aug 28, 2006. doi: 10.3748/wjg.v12.i32.5148
Revised: April 20, 2006
Accepted: April 24, 2006
Published online: August 28, 2006
AIM: To assess the feasibility of using BRAF, K-ras and BAT26 genes as stool-based molecular markers for detection of colorectal adenomas and hyperplastic polyps (HPs).
METHODS: We applied PCR-SSCP and direct sequencing to detect BRAF mutations of polyps and paired stool samples. Primer-mediated restriction fragment length polymorphism (RFLP) analysis and mutant-enriched PCR were used in detection of K-ras mutations of polyp tissues and paired stool samples respectively. BAT26, a microsatellite instability marker was examined by detection of small unstable alleles in a poly (A) repeat.
RESULTS: No genetic alterations were detected in the 36 colonoscopically normal patients in either tissues or stools. BRAF, K-ras and BAT26 mutations were found in 4 (16%), 10 (40%) and 3 (12%) of 25 adenoma tissues and among them, 75%, 80% and 100% of patients were observed to contain the same mutations in their corresponding stool samples. In HPs, mutations of BRAF and K-ras were detected in the tumor DNA of 2 (11.1%) and 8 (33.3%) of 18 patients respectively, all of whom had identical alterations in their stools. Taken together, the three genetic markers detected 15 (60%) of 25 adenomas and 8 (44.4%) of 18 HPs. The sensitivity of stool detection was 80% for adenomas and 100% for HPs with an overall specificity of 92% for adenomas and 100% for HPs.
CONCLUSION: BRAF, K-ras and BAT26 genes have the potential to be molecular markers for colorectal adenomas and HPs, and can be used as non-invasive screening markers for colorectal polyps.
- Citation: Jin YM, Li BJ, Qu B, Du YJ. BRAF, K-ras and BAT26 mutations in colorectal polyps and stool. World J Gastroenterol 2006; 12(32): 5148-5152
- URL: https://www.wjgnet.com/1007-9327/full/v12/i32/5148.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i32.5148
Colorectal cancer (CRC) is one of the most common malignancies in the world and it is a disease that can be easily cured either by surgery or by endoscopic excision when diagnosed at an early stage[1]. It has been generally accepted that a majority of CRC develop through a well-defined adenoma-carcinoma sequence in which multiple genetic changes are involved in this pathway that is known as chromosomal instability pathway. K-ras gene is known to play an important role along this pathway in transitioning from early to intermediate adenomas[2]. However, increasing evidence accumulates that a subset of CRC arises via the hyperplastic polyp (HP)-serrated adenoma (SA)-carcinoma sequence that is associated closely with microsatellite instability (MSI) positive colorectal carcinomas[3-5]. These polyps are usually large and/or multiple and/or located in the proximal colon[6,7]. APC mutation is not involved in this pathway whereas K-ras and BRAF mutations are frequently observed in these polyps with MSI[8,9]. Both BRAF and K-ras are proto-oncogenes that interact in tandem in the RAS-RAF-MEK-ERK-MAP kinase signaling pathway, which plays an important role in the control of cell differentiation, proliferation, survival, and apoptosis[10].
HPs have long been regarded as safe lesions without neoplastic potential[11]. However, this view has been changed due to the discovery of the HP-SA-carcinoma sequence. Now it is believed that adenomas as well as some types of HPs are precursors of sporadic CRCs that may eventually develop into adenocarcinomas[2,5]. So detection and surveillance of these premalignant polyps may be of great importance in reducing the incidence of colorectal cancer. BRAF and K-ras mutations have been frequently observed in both adenomas and HPs[8-10]. This prompted us to detect colon polyps through analysis of these genes in stool DNA. In this study, we detected mutational activation of the K-ras and BRAF genes and along with an MSI marker, BAT26 in patients with adenomas or HPs, and performed a pair comparison between tumor tissue and stool sample in individual patients to assess the feasibility of using these genes as molecular markers for colon polyps.
We recruited 79 patients who underwent colonoscopy for various reasons at the 2nd Affiliated Hospital of Harbin Medical University from June 2004 to March 2005. These included 36 control patients without neoplasms, 18 patients with hyperplastic polyps and 25 patients with adenomas. Diagnosis was histologically confirmed. The mean age of control patients was 49.6 years compared with 51.6 years for patients with HPs and 56.5 years for patients with adenomas. Of the 43 polyps, 34 were from males and 9 were from females. Twenty-seven were from the left colon and 16 were from the right colon. The size of these polyps ranged from 3 to 50 mm in maximal dimension (mean = 5.28 mm for HPs and 18.7 mm for adenomas). Patients who had familial adenomatous polyposis or hereditary nonpolyposis colon cancer, and inflammatory bowel diseases were excluded.
Fresh stool specimens were collected from patients prior to colonoscopy. All the patients were given detailed oral and written instructions for stool collection. Stool samples were frozen immediately at -20°C after collection and transferred to -80°C for permanent storage within 24 h.
With the informed consent of all patients and approval of the ethics committee, paired bioptic polyp tissues were obtained during polypectomy. One part of the tumor was snap frozen in liquid nitrogen and stored at -80°C until the extraction of nucleic acids. Another part was fixed in formalin and paraffin-embedded for diagnosis.
DNA was isolated from stool samples by means of the QIAamp DNA stool mini kit (QIAGEN, USA) and from snap frozen tissues using Trizol reagent (Invitrogen, Life Technologies, USA) according to the manufacturer’s instructions.
To detect possible sequence alterations in BRAF, we performed nonisotopic single-strand conformational polymorphism (SSCP) analysis. The complete sequence of exon 15 of the BRAF gene was amplified from 50 ng of genomic DNA using the primers described previously[9]. PCR was carried out for 40 cycles with initial denaturation at 95°C for 5 min, followed by 95°C for 40 s, 55°C for 40 s, and 72°C for 40 s in a reaction volume of 25μL. The PCR products were then separated on 8% nondenaturing polyacrylamide gels (29:1) with or without 5% glycerol, and electrophoresis of the gels was carried out at 4°C-8°C.The gels were then fixed in 10% acetic acid, silver stained in a freshly prepared silver nitrate (0.1%) and developed in 3% sodium carbonate with 0.05% formaldehyde and 0.2% sodium thiosulphate.
All PCR products showing mobility shifts on SSCP were reamplified and purified, and evaluated by direct sequencing with an ABI 3700 DNA sequencer (Applied Biosystems).
The mutations at codons 12 and 13 of K-ras gene in tumor samples were screened by primer-mediated restriction fragment length polymorphism (RFLP) analysis as described[12]. Human placental DNA (Sigma, USA) was used as a wild type control. Colorectal carcinoma DNA with known mutations at K-ras codon 12 (GGT to GTT) and at codon 13 (GGC to GAC) was used as mutant controls.
Mutant-enriched PCR was used to analyze the mutational status of the K-ras gene in stool samples. The procedure was the same as described[12] with the only exception that 5 U of BstNI or BglI(NEB, Beijing) was used for the digestion, and the reaction was completed overnight. Final products were separated on 4% agarose gel and visualized by ethidium bromide staining. The same positive and negative controls were used as in PCR-RFLP analysis.
The MSI marker used was BAT-26, a mononucleotide repeat that by itself is a very good measure of generalized instability. PCR reactions were performed with the specific primers reported previously [13]. PCR products were loaded on 8% polyacrylamide/ 7 mol/L urea DNA-denaturing sequencing gels and silver stained as described above.
Differences between groups were assessed by × 2 test and Fisher’s exact test. All P values were two sided. Factors with P < 0.05 were considered statistically significant. The 95% confidence intervals (CI) were determined based on the exact binomial distribution.
Twenty-five sporadic adenomas and 18 hyperplastic polyps were collected and analyzed for the alterations of K-ras, BRAF and BAT26 genes. The median age of the patients with HPs was 56 years (range: 28-70) compared with 61.8 years for patients with adenomas (range: 40-74). Thirty-six stool samples from control patients were also detected.
Mutations of exon 15 of BRAF and codons 12, 13 of K-ras genes were analyzed, as these cover most of the mutation hot spots known of the two genes. The alterations of BRAF, K-ras and BAT26 genes in adenomas and their relationship with clinico-pathological characteristics are shown in Table 1.
| n | BRAFn (%) | K-rasn (%) | BAT26n (%) | All markersn (%) | |
| Adenomas | 25 | 4 (16) | 10 (40) | 3 (12) | 15 (60) |
| Gender | |||||
| M | 19 | 3 (15.8) | 7 (36.8) | 2 (10.5) | |
| F | 6 | 1 (16.7) | 3 (50) | 1 (16.7) | |
| Age (yr) | |||||
| < 60 | 14 | 3 (21.4) | 7 (50) | 3 (21.4) | |
| ≥ 60 | 11 | 1 (9.1) | 3 (27.3) | 0 | |
| Location | |||||
| Distal | 16 | 2 (12.5) | 8 (50) | 1 (6.3) | |
| Proximal | 9 | 2 (22.2) | 2 (22.2) | 2 (22.2) | |
| Size (mm) | |||||
| < 10 | 5 | 1 (20) | 2 (40) | 0 | |
| ≥ 10 | 20 | 3 (15) | 8 (40) | 3 (15) | |
| HPs | 18 | 2 (11.1) | 6 (33.3) | 0 | 8 (44.4) |
| Gender | |||||
| M | 15 | 2 (13.3) | 5 (33.3) | 0 | |
| F | 3 | 0 | 1 (33.3) | 0 | |
| Age (yr) | |||||
| < 60 | 12 | 2 (16.7) | 5 (41.7) | 0 | |
| ≥ 60 | 6 | 0 | 1 (16.7) | 0 | |
| Location | |||||
| Distal | 11 | 1 (9.1) | 4 (36.4) | 0 | |
| Proximal | 7 | 1 (14.3) | 2 (28.6) | 0 | |
| Size (mm) | 0 | ||||
| < 10 | 15 | 0 | 4 (26.7) | 0 | |
| ≥ 10 | 3 | 2 (66.7)1 | 2 (66.7) | 0 | |
| Number of HP | |||||
| SP | 4 | 1 (25) | 1 (25) | 0 | |
| MP | 14 | 1 (7.1) | 5 (35.5) | 0 | |
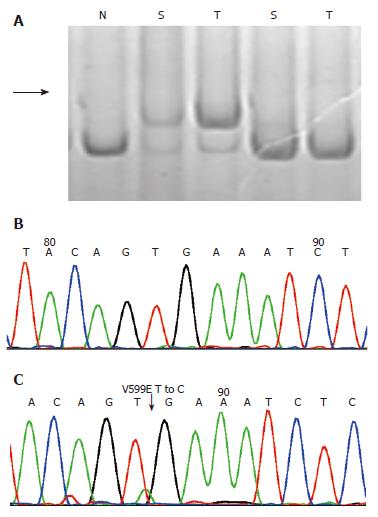
Of 25 tumor samples detected, 15 [60%, 95% CI: 39%-79%] were found to have at least one alteration in BRAF, K-ras or BAT26 genes. BRAF mutations were identified in 4 (16%, 95% CI: 5%-36%) cases, all at nucleotide position 1799 with T-A transversions (V599E) as confirmed by direct sequencing (Table 1, Figure 1). No significant correlation was found between BRAF mutations with patient’s gender, age, location of tumor and tumor size (Table 1). The identical V599E mutations of the BRAF gene were also observed in 3/25 (12%, 95% CI: 3%-31%) of fecal samples with a 75% agreement between tumor and stool.
In total, 3/25 (12%, CI: 3%-31%) of tumor DNAs showed BAT26 alterations (Table 1) and the same mutations were also observed in all the 3 paired stool samples. Two of the 3 samples with a BAT26 alteration also harbored a BRAF mutation that showed a close relationship between BRAF mutation and MSI (66.7% vs 4.5%, P < 0.05, Table 2). All 3 BAT26 positive samples were found in patients younger than 60 year and in adenomas larger than 10 mm in size but of no statistical significance. Also no significant relationship was found with respect to patient’s gender and tumor site.
| No of samples | No of BRAF mutation (%) | |
| Adenomas | 25 | 4 (16) |
| BAT26 (+) | 3 | 2 (66.7)1 |
| BAT26 (-) | 22 | 1 (4.5) |
| HPs | 18 | 2 (11.1) |
| BAT26 (+) | 0 | 0 |
| BAT26 (-) | 18 | 2 (11.1) |
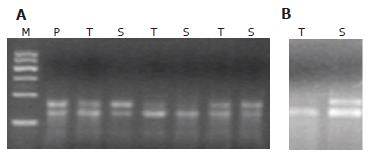
Among 25 tumor samples analyzed for K-ras mutations at codons 12 and 13, 10 (40%; 95% CI: 21%-61%) samples were found to carry a mutation, 8 of which were at codon 12 and 2 at codon 13 (Table 1, Figure 2). Mutations of the K-ras gene in corresponding stool samples were detected by mutant-enriched PCR[12]. Of 25 stool samples analyzed, K-ras alterations were observed in 9 (36%, 95% CI: 18%-58%) patients, 7 at codon 12 and 2 at codon 13. Eight of 10 stool DNAs had a mutation that matched exactly with the results of the corresponding tumor analysis (sensitivity: 80%, 95% CI: 44%-97%) whereas there was one case in which a K-ras mutation at codon 13 was observed in stool DNA but could not be detected in the corresponding tumor (specificity: 94%, 95% CI: 71%-100%, Figure 2). None of the tumor or stool samples with V599E carried a K-ras mutation.
To sum up, the three genetic markers detected 13/25 (52%) of stool samples of adenoma patients. The overall sensitivity of stool analysis using these three genes was 80% (12/15, 95% CI: 52%-96%) with a specificity of 92% (12/13, 95% CI: 64%-100%).
Eight (44.4%, 95% CI: 22%-69%) of the 18 HPs were found positive in either the BRAF or K-ras gene while no alterations were found in the BAT26 gene (Table 1). In corresponding stool samples, the same mutations were identified in all the 8 samples whose tumors contained the alterations with a 100% agreement between tumors and stools. No alterations were found in the other 10 samples. The specificity was also 100%. BRAF mutations were more common in polyps larger than 10 mm in size (P < 0.05, Table 1). BRAF and K-ras mutations were also mutually exclusive. No alterations were found in any of the three markers tested in 36 control DNAs of fecal origin.
In this study, we assessed the use of genetic alterations of BRAF, K-ras and BAT26 genes as stool-based molecular markers for colon adenomas. To our knowledge, this is the first time to show the results of stool-based gene detection of colorectal hyperplastic polyps.
We identified 16% of BRAF mutations and 40% of K-ras mutations in adenoma tissues. The results are similar to those reported previously[10]. However, in HPs, we observed a relatively lower incidence of BRAF mutations and a higher incidence of K-ras mutations than other studies [8,9]. Chan[8] reported a 36% BRAF mutation rate in HPs and Yang[9] reported an even higher mutation rate of 69.6%, but our result was only 11.1%. The incidence of K-ras mutations was 18% and 20% respectively by Chan[8] and Yang[9] but ours was 33.3%. This difference may be due to the small sample numbers in our study. The only form of BRAF mutation observed in our study was V599E. It is the most common mutation identified to date[8-10]. This missense mutation has been proven to maximally activate kinase activity of the BRAF protein and mutated forms of BRAF can transform NIH3T3 cells[14]. It is obvious that this variant has a strong functional selection for growth advantage. The high incidence of mutually exclusive mutations of BRAF and K-ras in both adenomas and HPs supports the previous hypothesis that activations of both genes are an early event in tumorigenesis of CRC[8-10].
BAT26 alteration was observed in 3/25 (12%) of adenomas, two of which also harbored a BRAF mutation. This close relationship between BRAF and MSI status agrees well with the findings in sporadic CRC[10]. By contrast, no BAT26 alteration was found in HPs. High level MSI was more often found in tumors located in the right colon, and most of our HPs were located in the left colon. However, several other studies have also reported a low MSI in HPs and serrated adenomas[15].
It is interesting to note that although not significant, in our study, patients with < 60 years of age had a tendency towards a lower prevalence of any mutation in both the adenoma and HPs groups. This may be due to the small sample numbers detected in our study, and larger sample analysis is needed to prove this phenomenon.
Based on our findings in the adenomas and HPs, we tried to use these three genes as stool based molecular markers for colon adenomas and HPs. Of the 15 adenomas that showed positive results, 12 (80%) samples have been detected to be positive in corresponding stool DNA, with a specificity of 92%. This result is quite comparable with the findings reported previously[16-20], in which DNA panel targeted mutations at K-ras, APC and p53 genes as well as BAT26 and long DNAs were used. But our detection seemed to be more convenient. First, mutation at the BRAF V599E hotspot is relatively simple to detect using SSCP and the sequencing method used in the present work (Figure 1). Second, the entire sequence of BRAF exon 15 is rather short, about 250 bp long, which can be amplified effortlessly, even in the stool DNA (data not shown).
In our study, we have detected a K-ras mutation in one stool DNA that could not be observed in corresponding adenoma tissue DNA. This is not caused by the different detection method we used in stools and tissues, because we analyzed this tissue DNA again using mutant-enriched PCR as in stool DNA, but still no mutation was found. This disparity might be due to other polyps present in the colon of this patient which we failed to detect through colonoscopy or because other neoplasms outside the colon were present in this patient that we did not know, since K-ras mutations have been detected in patients with pancreatic diseases[21].
In addition to adenomas, we also analyzed the DNA panel in patients with HPs. Our results showed that 44.4% of stool samples carried a mutation of either the BRAF or K-ras gene. All the mutations found in HPs could also be detected in corresponding stool DNA with a specificity of 100%. The results indicate that the mutations in HPs, even if they come from a very small tumor, can be detected in the stool DNA. It also proves that the technique of stool-based DNA detection for colorectal tumors is very sensitive.
Considering the small sample numbers and selected populations of symptomatic patients in our study, large investigations of fecal DNA analysis using these genes in asymptomatic populations are needed. In addition, recent investigations have shown that methylation of the hMLH1 is very common in HPs with MSI[5]. Thus, the combinations of genetic markers with epigenetic methylated genes will probably increase the sensitivity of the colorectal polyps detection rate.
In summary, our data indicate that mutations of BRAF, K-ras and BAT26 genes in adenomas and HPs are frequent and can be detected in corresponding stool samples. They can be used as stool based genetic markers for detection of colon polyps.
We thank Drs. Bing-Rong Liu, Shan-Ling Gao, Ming-Zi Han, Zhi-Wu Lu, Jing-Ming Guan, Feng-Hua Pei, Jing Chen and Xin-Hong Wang for their assistance with sample accrual; Thank Dr. Rei-Bo Zhao for pathological consultation; And thank Shu-Yun Zhang and Wei Liu for technical assistance and kindly providing us with the negative control DNA.
S- Editor Wang J L- Editor Zhu LH E- Editor Ma N
| 1. | Smith RA, Cokkinides V, Eyre HJ. American Cancer Society guidelines for the early detection of cancer, 2006. CA Cancer J Clin. 2006;56:11-25; quiz 49-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 310] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 2. | Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3455] [Cited by in RCA: 3345] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 3. | Hawkins NJ, Bariol C, Ward RL. The serrated neoplasia pathway. Pathology. 2002;34:548-555. [PubMed] |
| 4. | Jass JR, Whitehall VL, Young J, Leggett BA. Emerging concepts in colorectal neoplasia. Gastroenterology. 2002;123:862-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 323] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 5. | Hawkins NJ, Ward RL. Sporadic colorectal cancers with microsatellite instability and their possible origin in hyperplastic polyps and serrated adenomas. J Natl Cancer Inst. 2001;93:1307-1313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 220] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 6. | Jass JR. Serrated route to colorectal cancer: back street or super highway. J Pathol. 2001;193:283-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 7. | Higuchi T, Jass JR. My approach to serrated polyps of the colorectum. J Clin Pathol. 2004;57:682-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878-4881. [PubMed] |
| 9. | Yang S, Farraye FA, Mack C, Posnik O, O'Brien MJ. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol. 2004;28:1452-1459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 218] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 10. | Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 938] [Cited by in RCA: 952] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 11. | Williams GT. Metaplastic (hyperplastic) polyps of the large bowel: benign neoplasms after all. Gut. 1997;40:691-692. [PubMed] |
| 12. | Nollau P, Moser C, Weinland G, Wagener C. Detection of K-ras mutations in stools of patients with colorectal cancer by mutant-enriched PCR. Int J Cancer. 1996;66:332-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Rüschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749-4756. [PubMed] |
| 14. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7459] [Cited by in RCA: 7635] [Article Influence: 332.0] [Reference Citation Analysis (0)] |
| 15. | Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, Stephens P, Edkins S, Tsui WW, Chan AS. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451-6455. [PubMed] |
| 16. | Dong SM, Traverso G, Johnson C, Geng L, Favis R, Boynton K, Hibi K, Goodman SN, D'Allessio M, Paty P. Detecting colorectal cancer in stool with the use of multiple genetic targets. J Natl Cancer Inst. 2001;93:858-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 224] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 17. | Ahlquist DA, Skoletsky JE, Boynton KA, Harrington JJ, Mahoney DW, Pierceall WE, Thibodeau SN, Shuber AP. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology. 2000;119:1219-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 352] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 18. | Rengucci C, Maiolo P, Saragoni L, Zoli W, Amadori D, Calistri D. Multiple detection of genetic alterations in tumors and stool. Clin Cancer Res. 2001;7:590-593. [PubMed] |
| 19. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 525] [Article Influence: 25.0] [Reference Citation Analysis (3)] |
| 20. | Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 143] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 21. | Caldas C, Hahn SA, Hruban RH, Redston MS, Yeo CJ, Kern SE. Detection of K-ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res. 1994;54:3568-3573. [PubMed] |