INTRODUCTION
Liver regeneration requires various factors promoting cellular proliferation. Among these, augmenter of liver regeneration (ALR)[1], a novel cytokine, similar to the hepatic stimulator substance[2], has been shown to be mainly involved in the process of liver regeneration. ALR was cloned by Hagiya et al[1] from the weanling rat liver in 1994. Subsequently, Giorda et al[3] and Yang et al[4] cloned the cDNA of human ALR. It was found that rat, mouse, and human ALR genes (and protein products) are highly conserved, homologous and preferentially expressed in testis and liver[3]. ALR and the Saccharomyces cerevisiae homologue Erv 1p are members of the new ALR/Erv 1 protein family[5-7]. Experiments and clinical research demonstrated that ALR could specifically promote hepatocyte proliferation and rescue liver failure[8-13]. The specific molecular impact of ALR on liver regeneration still remains unclear. Wang et al[14] have identified and characterized the receptor for human ALR on rat hepatocytes, and they found that human hepatoma cells via their specific receptor ALR may initiate a corresponding cytoplamic signal transduction. But it was also reported that ALR promotes liver regeneration by inhibiting the activity of hepatic natural killer (NK) cells[13] and rat ALR (rALR) has no effect on primary rat hepatocytes in vitro[15], suggesting that ALR may likely exert its biological effect by using hepatic non-parenchymal cells. Kupffer cells, the resident liver macrophages, have been implicated as the primary source of inflammatory factors but may also be the source of important growth factors and cytokines that initiate cellular recovery and regeneration[16,17]. The protective role of Kupffer cells in hepatic injury by promoting liver regeneration has been recently emphasized[18-20], but the relationship between ALR and Kupffer cells in liver regeneration has not been investigated. Here we report the existence of ALR receptor in Kupffer cells and show the fact that ALR promotes hepatocyte proliferation induced by Kupffer cells.
MATERIALS AND METHODS
Isolation and primary culture of rat hepatocytes
Hepatocytes were isolated from male Sprague-Dawley rats (weighing 180-250 g, provided by Animal Center of Chinese Academy of Sciences) by in situ perfusion of the liver with collagenase (0.5 g/L type I CLSI, Sigma) as described previously[21]. The viability of the cells was determined by trypan blue exclusion. Preparations with their viability greater than 85% or higher were used. The cells were suspended in Williams’ medium E (Sigma) containing 100 mL/L fetal calf serum, 2 mmol/L L-glutamine, 7 mg/L insulin and 10 mmol/L HEPES, plated in 24-well plates (0.125 × 106 cells/ well), and incubated in an atmosphere of 95% air and 5% CO2 at 37°C. The medium was renewed after incubated for 3 h and cells were used for experiments after an overnight incubation.
Preparation of Kupffer cells
Kupffer cells were prepared from the livers of male Sprague-Dawley rats (200-224 g) as described previously[22]. Briefly, following the collagenase (type IV from 0.25 g/L clostridum histoticum, Sigma)/protease (type XIV from 0.5 g/L streptomyces griseus, Sigma) digestion of liver and removal of hepatocytes and cell debris by low speed centrifugation, Kupffer cells and endothelial cells were purified from other non-parenchymal cells by density gradient centrifugation using 300 g/L metrizamide. Finally Kupffer cells were isolated by a centrifugal elutriation procedure[22]. The viability of Kupffer cell preparation was greater than 95% as determined by trypan blue exclusion. Kupffer cells were suspended in Williams’ medium E containing 100 mL/L heat-inactivated fetal calf serum, 5 MU/L penicillin and 5 g/L streptomycin and plated in 24-well culture plates at a density of 1 × 106 cells/well, then plated in 48-well culture plates at a density of 0.5 ×106 cells/well and incubated in an atmosphere of 95% air and 5% CO2 at 37°C. The medium was renewed after incubated for 3 h and cells were used for experiments after an overnight incubation.
Determination of the effects of Kupffer cells-conditioned medium on hepatocytes
Kupffer cells were washed and placed in serum-free Williams’ medium E containing 0.1% bovine serum albumin (BSA, Sigma) and the test ALR concentrations (control, 1, 10, 100 and 1000 μg/L). After 24 h, the medium was filtered (0.2 μm, Gibco) and transferred to the cultured hepatocytes. Hepatocytes were incubated for 24 h with the test agents, the medium was conditioned by Kupffer cells, and various determinations were then made.
3H-thymidine incorporation assay: Determination of DNA replication
Kupffer cells or hepatocytes were rinsed twice with Hanks’ buffered saline solution (HBSS), and placed in serum-free Williams’ medium E containing 1 g/L BSA and the test ALR concentrations (control, 1, 10, 100 and 1000 μg/L) for 24 h. After the medium was aspirated from Kupffer cells or hepatocytes, a fresh medium containing 37 MBq/L 3H-thymidine (Amersham Pharmacia Biotech, USA) was added to the culture wells. Following an incubation of 4 h at 37°C, the cells were washed with ice-cold HBSS containing 0.1% BSA, treated with 10% ice-cold trichloroacetic acid (TCA) for 10 min, and washed once with TCA followed by 95% ethanol. The cells were then digested with 5% sodium dodecyl sulfate (SDS) at 60°C for 25 min. The liquid was transferred into scintillation vial, and washed once more with 0.5 mL 0.5% SDS. The radioactivity was determined using 5 mL β-scintillation solution.
3H-leucine incorporation assay: Determination of protein synthesis
Kupffer cells or hepatocytes were stimulated for 24 h as in the 3H-thymidine incorporation assay. After the medium was aspirated, a fresh medium containing 37 MBq/L 3H-leucine (Amersham Pharmacia Biotech, USA) was added to the culture wells. Following an incubation of 4 h at 37°C, the medium aspirated from Kupffer cells or hepatocytes was transferred to 1.5 mL Eppendorf tubes, BSA (finally 5%) and TCA (finally 5%) were added for 15 min, centrifuged at 12 900 r/min for 20 min at 4°C, washed three times with 5% ice-cold TCA. Then 200 μL 0.1mol/L NaOH was added to dissolve the pellet, pH was adjusted with 0.1 mol/L HCl, the liquid was transferred to the scintillation vial. The cells were washed with ice-cold HBSS containing 1 g/L BSA, treated with 100 g/L ice-cold TCA for 10 min, and washed once with TCA followed by 95% ethanol. The cells were then digested with 5% SDS at 60°C for 25 min. The liquid was transferred to scintillation vial, and washed once with 0.5 mL 0.5% SDS, the radioactivity was determined using 5 mL β-scintillation solution.
BrdU incorporation assay
Cells were plated in 12-well plates, serum-starved, and stimulated as described previously in the 3H-thymidine incorporation assay. After 24 h of stimulation, the cultures were incubated with BrdU labeling reagent (Dako) diluted at 1:1000. Cells were rinsed and fixed for 30 min at room temperature with acetic alcohol (90% ethanol, 50 mL/L acetic acid, 50 mL/L distilled H2O). Endogenous peroxidase was blocked by incubation in 1% H2O2 in methanol for 20 min at room temperature. Cells were washed in PBS and incubated for 15 min in 1.5 mol/L HCl at 37°C, and extensively washed in PBS. Cells were then incubated in anti-BrdU antibody (mouse IgG, Sigma) diluted at 1:40 in PBS containing 1% BSA for 1 h at 37°C and washed in PBS. Cells were incubated in anti-mouse secondary antibody (goat anti-mouse IgG, Sigma) diluted at 1:100 in PBS, and 10 g/L BSA for 30 min at room temperature followed by washing. Cells were incubated in ABC solutions for 30 min at room temperature, washed and incubated in 50 mmol/L Tris (pH 7.6) for 5 min, then the 3, 3-diaminobenzidine solution was added. Three areas in each well were counted for a total of about 1000 cells. Proliferation was indicated as a percentage of labeled nuclei.
125I- recombinant rat ALR binding to cultured kupffer cells and primary hepatocytes
Recombinant rat ALR (rrALR) was expressed in Escherichia coli and prepared with high purity (> 95%) as described previously[23]. rrALR was radioiodinated with the chloramines-T methods[14,24]. Briefly, 15 uL of 50 mmol/L sodium phosphate buffers (pH7.0) and 18.5 MBq/L sodium -125I (Amersham Pharmacia Biotech, USA) were added to a siliconized tube containing 5 μg of rrALR. The reaction was started by adding 10 μL of chloramine -T reaction using 20 μL of ending solution (50 nmol/L N-acetyl-L-tyrosine, 0.01 mol/L sodium phosphate buffer). 125I-rrALR was separated from free iodine by gel filtration on a column (20 cm× 11.0 cm) of Sephadex G-25 (Amersham Pharmacia Biotech, USA) equilibrated with PBS and 1 g/L BSA. The fractions containing 125I-rrALR were pooled. The assay was performed as described previously[14,23]. Kupffer cells and rat hepatocytes were washed in HBSS (containing 20 mmol/L HEPES, 1.3 mmol/L CaCl2, 0.5 mmol/L MgCl2, 1% BSA, pH 7.0), and pre-incubated in the presence of the same buffer for 30 min at 25°C. After equilibration was achieved in this medium, 0.3 mL containing 12.5 pmol/L-3200 pmol/L 125I-rrALR with or without 5 μmol/L unlabeled rrALR( saturation binding) was incubated for 3 h at 25°C with constant shaking. The cells were washed 4 times with ice-cold PBS and digested with 0.5 mL 0.75 mol/L NaOH for 30 min at 37°C, and the radioactivity in the digest was determined in a gamma-counter (cpm). Specific binding of 125I-rrALR was calculated as the difference between cell-associated radioactivity in the presence and absence of unlabeled rrALR.
Statistical analysis
The values are presented as mean ± SD of triplicate determinations. Each experiment was repeated at least three times. Student’s t-test was employed for statistical comparison of the paired samples. P < 0.05 was considered statistically significant.
RESULTS
Effects of ALR concentration on Kupffer cells
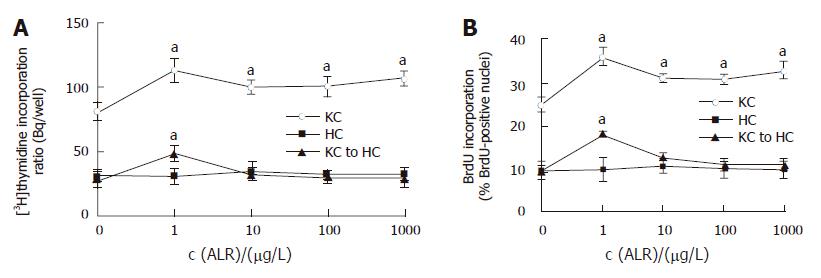
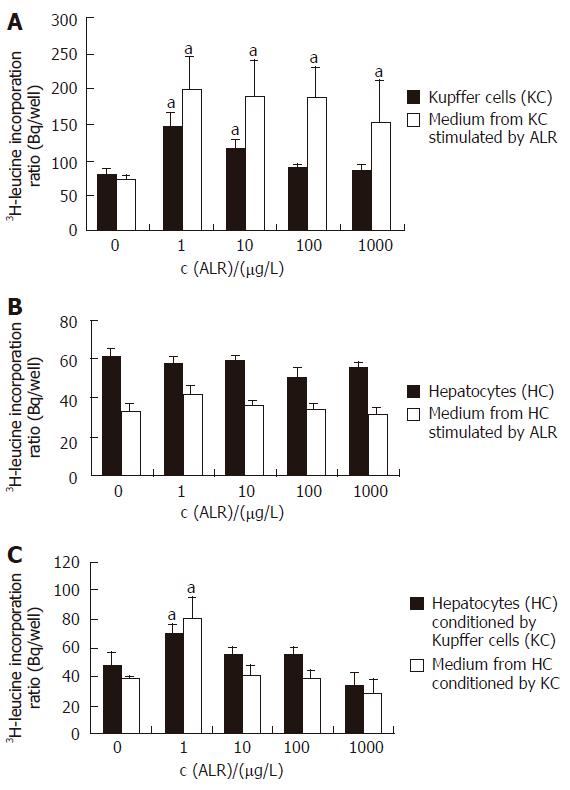
3H-thymidine incorporation assay showed that rrALR stimulated DNA replication in Kupffer cells in a non-concentration-dependent manner (Figure 1A). The significant effect was observed at the concentration of 1 μg/L ALR, the DNA replication increased to 144.2% as compared to the control (117 ± 11.7 Bq/well vs 81.2 ± 6.7 Bq/well, P < 0.05). Following the increase of added ALR concentration, DNA replication began to decrease, but was still higher than that in the control (P < 0.05). Being consistent with the 3H-thymidine incorporation assay, cell labeling by BrdU increased to 36% from a basal level of 24% at the concentration of 1 μg/L ALR (P < 0.05, Figure 1B, Figure 2). Similar to the DNA replication, ALR also produced a non-concentration-dependent transforming of protein synthesis both in Kupffer cells and in associated medium, but the significant effect was observed at the lowest concentration of 1 μg/L ALR tested (Figure 3A). The effects in cells and associated medium elevated to 182.6% and 275.3%, respectively, as compared to those in the control (cells:147.5 ± 19.6 Bq/ well vs 80.8 ± 5.9 Bq /well, P < 0.05; associated medium: 198.0 ± 46.1 Bq / well vs 71.9 ± 7.5 Bq/well, P < 0.05). Following the increase of added ALR concentration, protein synthesis in associated medium decreased, but was still higher than that in the control (P < 0.05, Figure 3A).
Figure 1 Effects of ALR on proliferation of Kupffer cells and hepatocytes.
A: DNA replication; B: BrdU-positive nuclei. KC: Kupffer cells; HC: Hepatocytes; KC to HC: Hepatocytes conditioned by Kupffer cells. aP < 0.05 vs control, n = 3.
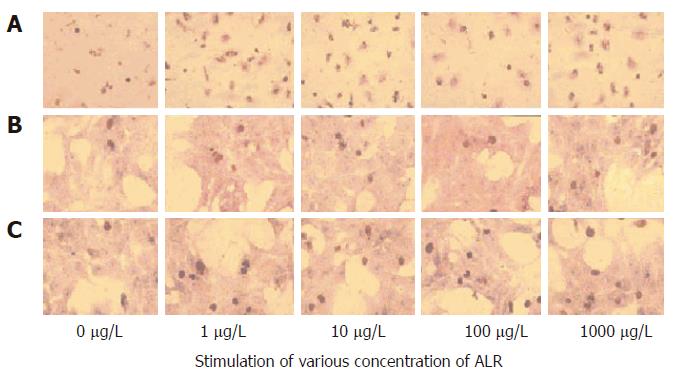
Figure 2 BrdU immunohistochemistry.
The nuclei of BrdU-positive cells were stained brown. A: Kupffer cells; B: Hepatocytes; C: Hepatocytes conditioned by Kupffer cells (× 100).
Figure 3 3H-leucine incorporation assay.
A: Kupffer cells and associated medium; B: Hepatocytes and associated medium; C: Hepatocytes conditioned by Kupffer cells and associated medium. aP < 0.05 vs control, n = 3.
Effects of ALR concentration on hepatocytes
ALR did not cause any significant change in DNA replication and protein synthesis in hepatocytes or in associated medium (Figures 1A and B, Figure 2, Figure 3B).
Effects of the medium conditioned by Kupffer cells with ALR on hepatocyte proliferation
Stimulation of hepatocytes with the medium conditioned by Kupffer cells with ALR increased DNA replication and protein synthesis at the lowest concentration of 1 μg/L ALR tested, to 174.3% (3H-thymidine incorporation, Figure1A) or 180% (BrdU lableling, Figure1B, Figure 2), 146.2% (in cells) and 210.7% (in associated medium) (Figure 3C) respectively, as compared to that in the control (DNA replication: 48.4 ± 3.5 Bq/ well vs 27.8 ± 4.7Bq/well in 3H-thymidine incorporation, 18.0% vs !0.0% in BrdU lableling; protein synthesis:70.5 ± 5.7 Bq/well vs 48.2 ± 9.2 Bq/well in cells, 80.7 ± 15.0 Bq/well vs 38.3 ± 2.5 Bq/well in associated medium, P < 0.05). But with the increase of added ALR concentration, the medium conditioned by Kupffer cells with ALR produced a concentration-dependent inhibition of DNA or protein synthesis in hepatocytes, the significant effect was observed at the highest concentration of 1000 μg/L ALR tested (DNA: 28.9 ± 5.8 Bq /well, Figure 1A; protein synthesis: 33.9 ± 9.0 Bq/well in cells, 29.3 ± 8.9 Bq /well in the medium, Figure 3C).
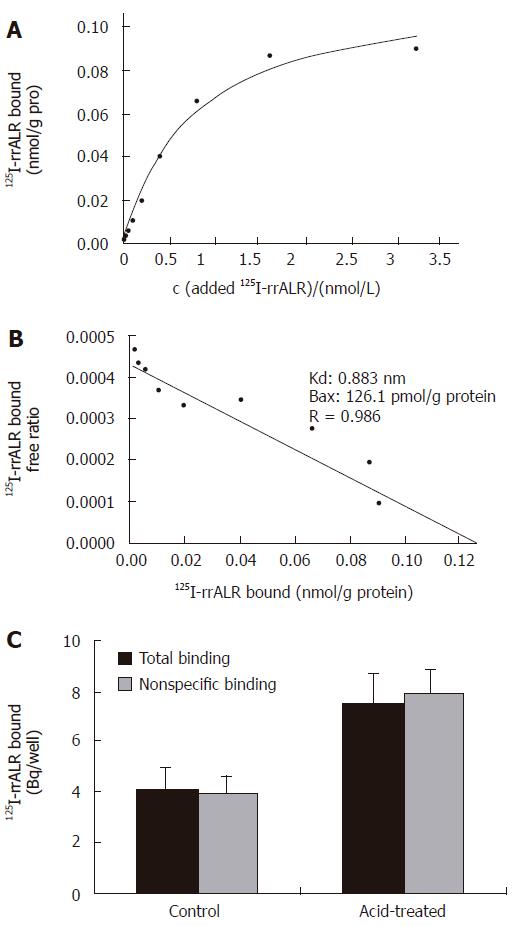
125I-rrALR binding assay
Typical saturation curves of 125I-rrALR binding to cultured Kupffer cells are shown in Figure 4A. Specific binding of ALR was saturated at about 1.5 nmol/L. Scatchard analysis resulted in a rectilinear plot (Figure 4B), thereby suggesting the presence of a single class of high affinity binding sites, namely the existence of a receptor of ALR on Kupffer cells. The 125I-rrALR binding capacity to cultured Kupffer cells was 126.1 ± 22.3 pmol/g protein, the affinity of the receptor was 0.883 ± 0.056 nmol/L.
Figure 4 125I-rrALR binding assay.
A: Saturation cures of 125I-rrALR binding to its receptor on Kupffer cells; B: Scatchard plot of the ALR receptor on Kupffer cells; C: Binding of ALR to cultured hepatocytes (mean ± SD, n = 3).
However, similar to Gandhi et al[23], no receptor for ALR was found on hepatocytes by radioligand binding analysis. To investigate the possibility that ALR receptors on hepatocytes are down-regulated, the acidic medium, a procedure known to dissociate bound peptide ligand from its receptors, was used to treat cultured hepatocytes as previously described[21,22], but it did not unmask any specific binding of ALR yet (Figure 4C).
DISCUSSION
Previous studies have revealed that ALR appears to be an important regulator of liver regeneration. Recently, emphasis has been placed on ALR molecular mechanisms underlying liver regeneration. Many advances have been gained in murine and human experiments. For instance, Yang et al[4] have cloned the cDNA of human ALR (hALR) from human fetal liver lysates encoding 125 amino acids and 15KD in molecular weight. hALR could stimulate proliferation of cultured hepatocytes as well as hepatoma cells in vitro and rescue acute hepatic failure in vivo. ALR exerts its hepatrophic activities through paracrine and autocrine pathways. Wang et al[14] have demonstrated the existence of hALR specific receptor on the surface of primary hepatocytes and hepatoma cells. Through the ALR receptor, ALR stimulates hepatocyte proliferation by activating the mitogen-activated protein kinase cascade (MAPK signaling pathway)[25]. The existence of ALR in nuclei and cytosol of liver tissues further implicates that ALR has intracellular function in hepatocytes[26]. In immunohistochemical studies, Thasler et al[27] found that hALR is mainly expressed both in normal and in impaired hepatocytes. Even though there exist many inconsistent issues, Gandhi et al[23] have failed to show any specific rALR receptor in hepatocytes. Hagiya et al[1] and Yu
et al[12] reported that rat ALR has no effect on primary rat hepatocytes in vitro and that the expression of ALR is absent in normal hepatic tissues[15]. The existence of contrary results together with the mechanism of inhibiting hepatic NK activity to promote liver regeneration suggests that it is possible for ALR to exert its bioactivities by interacting with other cytokines or with hepatic non-parenchymal cells on hepatocytes.
In this study, we showed the presence of high affinity receptors of ALR on hepatic Kupffer cells, and found that ALR could stimulate proliferation of Kupffer cells, suggesting that ALR stimulates hepatocyte proliferation by activating Kupffer cells, which may improve our understanding of the molecular mechanism of the biological action of ALR and its effect on liver regeneration. 3H-thymidine and 3H-leucine incorporation assay and BrdU labeling demonstrated that ALR stimulated DNA and protein synthesis of Kupffer cells in a non-concentration-dependent manner and had no effect on primary hepatocytes. But with the medium conditioned by Kupffer cells, ALR promoted hepatocyte proliferation. Similar to this study, Chen et al[28] reported that when hepatocytes are co-cultured with Kupffer cell supernatant only activated by LPS, hepatocyte DNA and protein synthesis are increased significantly. Recently Armburst et al[19] reported that the early gene expression of hepatocyte growth factor in Kupffer cells after carbon tetrachloride (CCl4) treatment may be an important event in the early phase of liver regeneration. Together with the finding about the entry of ALR into Kupffer cells by immunolabelling[23], these events suggest that the medium containing some kind of hepatocyte growth factors coming from Kupffer cells conditioned by ALR and secreting into the medium, can regulate hepatocyte proliferation. The increased 3H-leucine incorporation showed the protein synthesis both in Kupffer cells and in associated medium. What kind of protein is needed should be further studied. The foregoing studies have tested acute reactant cytokines such as interleukin-6 (IL-6) and tumor necrosis factor (TNF)-α originated mainly from activated Kupffer cells which are the chief growth factors in hepatocyte proliferation in vivo[17,29-33]. Since TNF-α and IL-6 are involved in the initiation of liver regeneration, we assume that ALR, like LPS, may activate Kupffer cells by unknown means and then mediate Kupffer cell-dependent release of TNF-α and IL-6, triggering hepatocyte proliferation. The ALR receptor on Kupffer cells can elucidate the effect of ALR on Kupffer cells in part. Here, the high affinity receptor for ALR on Kupffer cells can be identified by 125I-ALR binding assay. When ALR binds to the cell surface receptor, Kupffer cells are activated and then release various cytokines initiating liver regeneration. The binding effect is saturable. When an excess amount of unlabeled ALR is added, it could replace the cell surface sites to which ALR has bound. It was reported that 125I-ALR is found to have the similar molecular weight and biological activity as the unlabeled ALR, and that iodination does not change the characteristics of natural ALR, suggesting that 125I-ALR can be used for identification of ALR receptor[14]. As for the specificity of this binding, further researches need to be done. In our study, as the concentration of ALR was added to 1000 μg/L, the effect of ALR-dependent-Kupffer cells on hepatocyte proliferation was inhibited, suggesting that Kupffer cells have a dual function as a regulator in liver regeneration. Responding to the immoderate proliferation effect of hepatocytes, Kupffer cells exert their inhibitory effects by releasing growth inhibiting factors. Transforming growth factor(TGF)-β is the most obvious candidate because it has been shown in vivo models that TGF-β could antagonize TNF-α actions and induce apoptosis, constraining the amount of hepatocyte proliferation[34-36].
Similar to the study by Gandhi et al[23], no receptor for rALR was found on hepatocytes in our study, which may interpret the inability of rALR to exert a mitogenic effect on hepatocytes in vitro. To investigate the possibility that ALR receptors on hepatocytes are down-regulated, the acidic medium, a procedure known to dissociate bound peptide ligand from its receptors was used to treat cultured hepatocytes, but it did not unmask any specific binding of ALR. At the same time, there is evidence that hepatocytes synthesize and secrete ALR[23,37]. A further finding showed that hepatic ALR levels decreased while circulating ALR levels increased 12 h after 70% hepatectomy, suggesting that the release of stored ALR from hepatocytes rather than the accelerated synthesis increases the circulating ALR level[28].
How should these events be put together? We hypothesize that under pathological or biological circumstances of liver regeneration such as partial hepatectomy or weaning livers, activated Kupffer cells release various cytokines initiating liver regeneration, and that the receptor of ALR on hepatocytes (assuming its existence) and ALR which are constitutively expressed and stored in hepatocytes in an inactive form beforehand, are released from the cells in an active form by unknown means, then ALR exerts its hepatrophic activities through paracrine and autocrine pathways such as binding to Kupffer cells or the inhibition of hepatic NK cells to promote hepatocyte proliferation. To prevent excess hepatocyte proliferation, anti-hepatotrophic growth factors such as TGF-β are produced.
In conclusion, the mechanisms of liver regeneration are involved in complicated interactions of various cytokines and cells[29,37]. Kupffer cells contain high affinity receptors for ALR on cell surfaces, by which ALR promotes hepatocyte proliferation. Further studies focusing on the examination of the specificity of receptors on Kupffer cells are needed.