Published online Jul 21, 2006. doi: 10.3748/wjg.v12.i27.4401
Revised: April 28, 2004
Accepted: August 3, 2004
Published online: July 21, 2006
AIM: To construct the expression vectors for prokaryotic and eukaryotic human augmenter of liver regeneration (hALR) and to study their biological activity.
METHODS: hALRcDNA clone was obtained from plasmid pGEM-T-hALR, and cDNA was subcloned into the prokatyotic expression vector pGEX-4T-2. The recombinant vector and pGEX-4T-2hALR were identified by enzyme digestion and DNA sequencing and transformed into E coli JM109. The positively selected clone was induced by the expression of GST-hALR fusion protein with IPTG, then the fusion protein was purified by glutathine s-transferase (GST) sepharose 4B affinity chromatography, cleaved by thrombin and the hALR monomer was obtained and detected by measuring H thymidine incorporation.
RESULTS: The product of PCR from plasmid pGEM-T-hALR was examined by 1.5% sepharose electrophoresis. The specific strap was coincident with the theoretical one. The sequence was accurate and pGEX-4T-hALP digested by enzymes was coincident with the theoretical one. The sequence was accurate and the fragment was inserted in the positive direction. The recombinant vector was transformed into E coli JM109. SDS-PAGE proved that the induced expressive fusion protein showed a single band with a molecular weight of 41 kDa. The product was purified and cleaved. The molecular weights of GST and hALR were 26 kDa, 15 kDa respectively. The recombinant fusion protein accounted for 31% of the total soluble protein of bacterial lysate. HALR added to the culture medium of adult rat hepatocytes in primary culture and HepG2 cell line could significantly enhance the rate of DNA synthesis compared to the relevant control groups (P < 0.01).
CONCLUSION: Purified hALR has the ability to stimulate DNA synthesis of adult rat hepatocytes in primary culture and HepG2 cells in vitro, and can provide evidence for its clinical application.
- Citation: Zhang YD, Zhou J, Zhao JF, Peng J, Liu XD, Liu XS, Jia ZM. Expression, purification and bioactivity of human augmenter of liver regeneration. World J Gastroenterol 2006; 12(27): 4401-4405
- URL: https://www.wjgnet.com/1007-9327/full/v12/i27/4401.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i27.4401
Transplantation of hepatocytes can be used to treat acute and chronic hepatic failure, and related hepatic diseases[1,2]. Due to the difficulty to get satisfactory proliferation of transplanted cells, the hepatic function and superseding effect of the damaged hepatic parenchyma are not good[3,4]. A large number of living hepatocytes are needed in the substituting period for metabolic disorder[5]. But the source of donor hepatocytes is limited. All of above-mentioned factors influence hepatic transplantation.
Human augmenter of liver regeneration (hALR) is a stimulator of hepatic proliferation found in recent years. Compared to other growth factors, it can be used to restrain the growth of some tumor cells[6,7]. Meanwhile, it can resist hepatic damage and reverse hepatic fibrosis[8,9]. Application of this factor will improve the proliferating ability of culture in vitro and hepatocyte transplantation in vivo[10].
Plasmid pGEX-7-hALR containing 380 bp human regeneration promoter cDNA, prokaryote expression vector pGEX-4T-2 and host bacteria JM109 were conserved in our laboratory. Ligase T4 DNA, restrictive endonucleases BamH1 and EcoR1, calf intestinal alkaline phosphatase (CIP) and thrombin were obtained from American MBI Company. TaqDNA polymer dNTP was purchased from Promega Company. Standard proteins with a low molecular weight and calibration kit for SDS electrophoresis were purchased from Pharmacia Company MBI QIA-GEN gel callback sample box and emitting isotope H-TDR were bought from Shanghai Shenggong. Wistar male rats were provided by the Animal Center of our school.
In this research, recombinant hALRcDNA was cloned into plasmid pGEX-4T-2 with genes of fusion protein, and then recombinant vector was transformed into E coli JM109. After digestion and sequencing, fusion protein expression was detected by SDS-PAGE electrophoresis. Affinity chromatography was used to purify GST-hALR after digestion by thrombin. After hALR was cloned into an expression plasmid of eukaryon with expressive green fluorescence protein (EGFP) as reporter gene, LO2 hepatocytes were transfected by liposome transfection method. Transfection efficiency and instantaneous expression were observed after recombination. The biological activity of hALR was studied by detecting DNA synthetic rate as previously described[11].
Oligoneucleotide primer: According to the sequence of ALR, the specific primer of hALRcDNA was designed and the sequence identified by enzymes BamHI and EcoRI was used to observe and reverse the primer. The upper primer p1 was 5’-CGGGATATGGGGACGCAGCAGAAGAAGCGGGAC-3’, the lower primer p2 was 5’-GCGAATTCCGCTAGTCACAGGAGCCGACCTTCC-3’. After 35 cycles of amplification at 95°C for 1 min, at 64°C for 1min, at 72°C for 1min, 1.5% agarose gel eletrophoresis was used to identify the purified genes.
DNA recombination and identification: Gene expre-ssion sequence of hALRcDNA was cloned into pGEX-4T-2 vector, and identified with enzymes BamHI and EcoRI. The results were proved by Shanghai Physiologic Center.
Expression and purification of recombinant gene induced by eukaryon: Recombinant positive clone con-taining pGEX-4T-2-hALR was put into LB culture medium at 37°C, and IPTG (final concentration was 1 mmol/L) was added to induce expression for 4 h, then the bacilli were collected and broken by ultrasound pulverizer (4°C, 12 000 r/min, 20 min). The supernatant was collected and put into 2 × loading buffer. The expression of recombinant fusion protein was identified by SDS-PAGE eletrophoresis.
Recombinant plasmid transfection of LO2 hepato-cytes: LO2 hepatocytes were transfered into a cell culture flask in DMEM containing penicillin 100 U/mm, streptomycin 100 U/mm, 10% calf serum, tri-distilled water and incubated in 5% CO2 at 37°C.
One day before liposome transfection, LO2 hepatocytes 4 × 104 were loaded onto coverslips, put into 6-well plates and cultured with 1.5 mL DMEM containing 10% calf serum 5% CO2 at 37°C until cells grew to 80% coverage. The cells were washed 2 times with normal saline. DNA-liposome complex was put into two six-well plates, 0.5 mL of DMEM-SF without antibiotics was added into each well and the plates were slightly shaken to make the DNA-liposome mixture flow evenly over the cell surface. Cells were cultured in 5% CO2 at 37°C for 5 h without change of supernatant, 0.5 mL of 30% of calf serum was added into DMEM and cultured for 24 h, then the medium was washed two times with normal saline, 1.5 mL of 10% of serum was added into DMEM.
Instantaneous expression of recombinant plasmid in LO2 hepatocytes: LO2 hepatocytes were cultured in different groups. The first group was vaccinated on coverslips after digestion by pancreatin, put into six-well plates and incubated. The cells were observed under fluorecent microscope after 12, 24, 48 h of transfection. The second group of hepatocytes was transfected with LO2 hepatocytes of recombinant plasmid in DMEM-SE medium. The trial group was transfected with blank material into blank plasmid, the upper clear liquid was drawn after one week of culture, then 30% of volume was added into routinely cultured HepG2 cancerous cell medium and vaccinated into 12-well plates 5 × 105/mL, the HepG2 cells were incubated in routine culture medium in control group. Luci thymidine was added into each well after cultured for 24 h. The cells were collected, washed repeatedly with PBS, the infiltrative membrane was dried by airing, and put into a scintillation bottle with scintillation liquid. DNA synthesis rate (CPM value) was detected by a scintillometer. The third group was cultured routinely for 72 h, G418 200 μg/mL of calf serum was added to screen cells. The liquid was changed every 3 d.
Detection of biological activity of expression hALR: The primary hepatocytes were isolated by hepatic perfusion. In brief, 2.5 mL of mixed hepatocyte suspension and 15 mL of Percoll were centrifuged at 88 r/min, for 13 min, then the cells were collected and counted using a optical microscope. The separated hepatocytes and HepG2 tumor cells were incubated respectively in cell culture plates, and then the supernatant was discarded. The third trial group was established by adding DMEM liquid separately containing hALR 5 ng/mL, 10 ng/mL, and 20 ng/mL.
Statistical Analysis: The data were dealt with by double factor variance analysis with SPSS 10.0 software package.
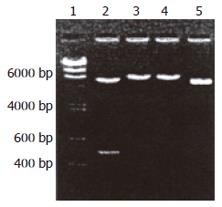
Construction and identification of pGEX-4T-2hALR: The reaction result of PCR augmentation of hALRcDNA (Figure 1) was coincident with hALRcDNA. There were the endonu-clease sites of BamHI, EcoRI on 5’ - 3’ ends.
Construction of pGEX-4T-2hALR after hALRcDNA acquired by PCR augmentation was performed using restriction endonucleases BamHI and EcoRI. Gene segment was recycled by 1.5% sepharose electrophoresis. PGEX-4T-2 was cleaved by BamHI and EcoRI, then the gene segment was recycled. Transformed susceptible bacteria JM109 were screened using ampycilline to yield 12 monoclone colonies. The recycled vector segment was not recombined and cultured directly in the contrast LB culture medium after phosphorylation, but no clone appeared. Five monoclone colonies were selected randomly. The plasmid was digested by BamHI and EcoRI to yield a band 5.3 kb (Figure 2). This indicated exogenous DNA was inserted. After digestion by BamHI and EcoRI and recombinant plasmid, two bands of 4.9 kb and 380 bp were produced. In contrast to non-vector double cutting enzyme, 380 bp external DNA was inserted in pGEX-4T-2 between the cutting site of BamHI and EcoRI. The recombinant plasmid-inserted 380 bp hALR was acquired. The structure of plasmid was completely satisfactory. Five positive clones were acquired. Therefore, a complete hRcDNA segment was inserted into pGEX-4T-2 plasmid between BamHI and EcoRI.
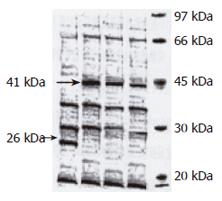
Expression of hALR in E coli: Expression of fusion protein GST-hALR was induced after the bacteria containing recombinant expression plasmid pGEX-4T-2hALR were induced by IPTG. The split product was detected by SDS-PAGE electrophoresis (Figure 3), a new band of 41 kDa was found in the recombinant bacteria, while the nonfusion protein GST band expressed by pGEX-4T-2hALR nonvector transformed bacteria was 26 kDa, indicating that molecular weight of the fusion protein was 41 kDa, the part of GST was 26 kDa, and the molecular weight of hALR was 15 kDa.
After the recombinant bacteria were split by ultro-sound, its inclusion body and supernatant were detected by SDS-PAGE electrophoresis (Figure 4). The significant band of fusion protein (GST-hALR) was seen at the site of protein 41 kDa in upper clean liquid, but the weak band was seen at the same site in inclusion body, indicating that the fusion protein was expressed mainly in the soluble form in the upper liquid.
To identify the best expression condition, the expression period was analyzed. The result showed that the expression product of fusion protein in the LB liquid culture medium was ascended in 0-8 h after the transformed bacteria were induced with IPTG under 30°C. By the gel scanning system, the expression amount of GST-hALR at 8 h covered 31% soluble protein of bacteria.
Fusion protein in the supernatant was analyzed by SDS-PAGE, then deoxidized glutathione S-transferase(GST) was used to purify GST-hALR. The vector of pGeX-4T-2 had the cutting site of the thrombin between GST and multicolone, the function of thrombin could separate GST and hALR in the fusion protein. After fusion protein was split by thrombin, split GDT and undigested fusion protein were filtered by glutathione sepharose 4B affinity chromatography. The purified hALR was acquired, and there were also single clean strips in the map of SDS-PAGE. Molecular weight was about 15 kDa. The concentration of the purified hALR was about 90.5% and the hALR protein concentration was about 0.87 mg/mL.
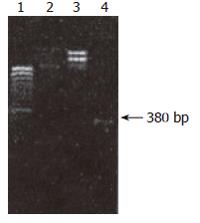
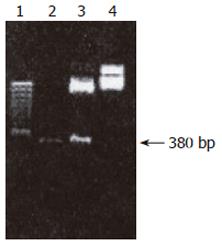
Construction and identification of recombinant plasmid pEGFP-C1-hALR: In the map of pEGFP-C1 multiple clones, EcoRI and BamI II were in the region of MCS. EcoRI was in the front, BamI II was in the rear, indicating that hALR could be inserted positively.pEGFP-C1 plasmid and ALRcDNA segment were cut respectively by EcoRI and BamI II. The site of cut hALR PCR segment was at 400 bp, two light bands were seen in the uncut pEGFP-C1 plasmid due to the difficult spatial conformation of cycled plasmid. pEGFP-C1 was cut to linear molecule by enzymes, a light band was seen by electrophoresis, the velocity rate was slightly faster than that of uncut pEGFP-C1 plasmid by enzymes (Figure 5).
When the kanamycin plate was incubated overnight at 37°C, 15 clones sprouted which were cultured overnight with kanamycin LB culture medium at 37°C.
The site of PCR augmetation band of recombinant plasmid was similar to that of PCR augmetation band of bank DNA. There were two characteristic bands after recombinant plasmid was cut by enzymes. One band was at 400 bp, the other band was at 4.7 kb, suggesting that hALRcDNA was cloned into pEGFP-C1. Recombinant plasmid pEGFP-C1-hALR was constructed successfully.
The result of PCR was coincident with the theoretical one, the sequence of recombinant plasmid pEGFP-C1-hALR was right.
OD value of pEGFP-C1-hALR detected by ultraviolet spectrophotometer is shown in Table 1.
| Sample | abs | abs | bkgabs | 260.0 nm | 280.0 nm | Protein | Nucleic acid |
| ID | 280.0 nm | 260.60 | 320.0 nm | 280.0 nm | 260.0 nm | μg/mL | μg/mL |
| 1 | 0.0319 | 0.0205 | 0.0057 | 1.7679 | 0.5657 | 3.1539 | 1.1124 |
| 0.0337 | 0.0234 | 0.0086 | 1.6953 | 0.5899 | 3.9632 | 1.0440 | |
| 2 | 0.0152 | 0.0073 | -0.0009 | 1.9486 | 0.5132 | 0.6326 | 0.7177 |
| 0.0159 | 0.0101 | -0.0009 | 1.8485 | 0.5410 | 1.6853 | 0.8892 |
pEGFP-C1-hALR and its sequence might be used to transfect cells and to observe the expression condition of green fluorescence protein.
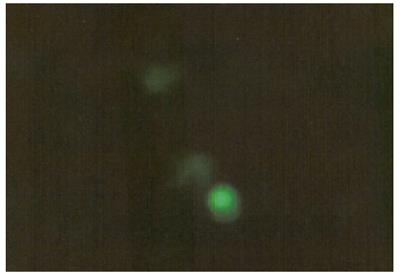
Expression of recombinant plasmid pEGFP-C1-hALR in LO2 hepatocytes: LO2 hepatocytes with recombinant plasmid pEGFP-C1-hALR for 12 h. One coverslip was taken from the 6-well plates transfected and observed under microscope. Cells were attached to the wall, cleavage phase of some cells were seen. The medium was washed 2 times with normal saline, digested by pancreatin. The cells were counted. Another coverslip was taken from the plates to observe the expression of green fluorescence under fluorescence microscope (Figure 6).
The green fluorescence was expressed in some cells after transfection of liposome for 12 h. Most of the expression was weak.
Detection of biological activity of hALR: The hepatocytes were separated by hepatic perfusion with IV collagenase. After separation and purification, the number of cells was about 2 × 108/mL, the survival rate of cells was about 94%. CPM value detected by 3H-thymidine incorporation method was used to observe the influence of DNA synthesis rate of hALR on primary cultures of hepatocytes and HepG2 cells cultured in vitro (Tables 2 and 3).
| CPM | 1 | 2 | 3 | 4 |
| Without hALRcontrol group | 5 ng/mL hALRmedium group | 10 ng/mL hALRmedium group | 20 ng/mL hALRmedium group | |
| n = 6 | n = 6 | n = 6 | n = 6 | |
| 1 1125 ± 158 | P < 0.01 | P < 0.01 | P < 0.01 | |
| 2 1802 ± 127 | P < 0.01 | P < 0.01 | ||
| 3 2764 ± 445 | P < 0.01 | |||
| 4 3344 ± 417 |
| CPM | 1 | 2 | 3 | 4 |
| Without hALRcontrol group | 5 ng/mL hALRmedium group | 10 ng/mL hALRmedium group | 20 ng/mL hALRmedium group | |
| n = 6 | n = 6 | n = 6 | n = 6 | |
| 1 1226 ± 158 | P < 0.01 | P < 0.01 | P < 0.01 | |
| 2 1802 ± 127 | P < 0.01 | P < 0.01 | ||
| 3 2764 ± 445 | P < 0.01 | |||
| 4 3344 ± 417 |
Seen from the above table, recombinant hALR might stimulate DNA synthesis of primary hepatocytes and HepG2 cells compared to control group. There was a significantly statistical difference (P < 0.01). The different dose of hALR had different effects on primary hepatocytes and HepG2 cells. Comparison of mean value between each group, there was a significant statistical difference (P < 0.01), indicating that DNA synthesis rate increased gradually with increasing dose of hALR.
Due to limited donor source and complication of the operation itself, liver transplantation in situ in treatment of terminal end hepatic disease has been widely used. The technique of hepatocyte transplantation developed at the end of the 1970's has gained more and more attention because one donor can provide hepatocytes for several recipients and the transplant technique is very simple[8,12].
We studied the biological activity of the expression product by constructing expression vector of pronuclei and eukaryons of human augmenter of liver regeneration in order to provide experimental evidence for the study of hepatocellular source of hALR hepatocyte transplantation modified by gene and the clinical application of hALR. But there are three major problems in the transplantation of hepatocytes, namely the transplanting location, immune reaction, and proliferation of hepatocytes[8]. When trans-planted in the parenchyma of spleen or in the abdo-minal cavity, hepatocyte transplantation always has better effects[13]. When biological materials such as biological compatibility membrane are used, hollow fibers may form a barrier for the transplanted hepatocytes, which has solved the problem of host immune repulsive action, and increased interest in xenogenic hepatocyte transplantation[11,14]. Transplanted hepatocytes perform the temporary maintenace of hepatic function and the temporary substitution for the damaged hepatocytes by proliferation in vivo. Since the desired results of culture in vitro and proliferation in vivo are difficult to get, the final result is not so good[3,4]. Furthermore, to treat congenial metabolic defects with hepatocyte transplantation, a large quantity of hepatocytes that can express exogenic genes are needed[15]. The traditional methods to improve the transforming rate such as 70% resection of liver, damaging the liver with CCL4, can bring damage to the liver of the recipient. Therefore how to acquire a stronger proliferation ability, how to culture plenty of cells in vitro, while ensure their biological activity and long lifespan are crucial problems.
With the advance in modern gene engineering techniques, especially combination of gene engineering techniques with modern surgical transplantation technique, a new curative technique is produced, namely genetically-modified cell transplantation, proliferation of the transplanted hepatocytes is possible to be solved through the modification of hepatocytes. HALR is a stimulator of hepatocyte proliferation that can bear temperature, with specificity for liver but without species specificity. HALR can accelerate synthesis of DNA of primary generative hepatocytes cultured in vitro and hepatogenous hepatic tumor cells and the proliferation of the cells, while it has no influence on the nonhepatic cells, showing that it has a specific role in accelerating proliferation of the hepatocytes. HALR is an important regeneration-regulating factor because it can accelerate the hepatocyte proliferation. HALR may accelerate proliferation of hepatic cells in 1/3 of models with partial resection of liver, indicating it is an important regeneration-regulating factor[11,16-18]. In addition, it can protect the liver against damage, reverse hepatic fibrosis[19,20] and take part in cell immune action. All these suggest that proliferation of the transplanted hepatocytes can be suppressed by the use of enhanced and modified regenerative cells. Regeneration-enhanced factor as a kind of special stimulating factor of growth of hepatic cells, has gained more and more attention of domestic and foreign scholars. It has a prospect to be used as an effective medicine to treat severe hepatic diseases.
S- Editor Wang J L- Editor Wang XL E- Editor Liu WF
| 1. | Kobayashi N, Miyazaki M, Fukaya K, Inoue Y, Sakaguchi M, Uemura T, Noguchi H, Kondo A, Tanaka N, Namba M. Transplantation of highly differentiated immortalized human hepatocytes to treat acute liver failure. Transplantation. 2000;69:202-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 55] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Nagata H, Ito M, Cai J, Edge AS, Platt JL, Fox IJ. Treatment of cirrhosis and liver failure in rats by hepatocyte xenotransplantation. Gastroenterology. 2003;124:422-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 89] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 3. | Cantz T, Zuckerman DM, Burda MR, Dandri M, Göricke B, Thalhammer S, Heckl WM, Manns MP, Petersen J, Ott M. Quantitative gene expression analysis reveals transition of fetal liver progenitor cells to mature hepatocytes after transplantation in uPA/RAG-2 mice. Am J Pathol. 2003;162:37-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Weglarz TC, Degen JL, Sandgren EP. Hepatocyte transplantation into diseased mouse liver. Kinetics of parenchymal repopulation and identification of the proliferative capacity of tetraploid and octaploid hepatocytes. Am J Pathol. 2000;157:1963-1974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 96] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Uyama S, Kaufmann PM, Kneser U, Fiegel HC, Pollok JM, Kluth D, Vacanti JP, Rogiers X. Hepatocyte transplantation using biodegradable matrices in ascorbic acid-deficient rats: comparison with heterotopically transplanted liver grafts. Transplantation. 2001;71:1226-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Tanigawa K, Sakaida I, Masuhara M, Hagiya M, Okita K. Augmenter of liver regeneration (ALR) may promote liver regeneration by reducing natural killer (NK) cell activity in human liver diseases. J Gastroenterol. 2000;35:112-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Zhang Y, Yang XM, Wang G, He FC. [Biological activity of recombinant human hepatopoietin]. Shengli Xuebao. 1999;51:347-350. [PubMed] |
| 8. | Kobayashi N, Ito M, Nakamura J, Cai J, Gao C, Hammel JM, Fox IJ. Hepatocyte transplantation in rats with decompensated cirrhosis. Hepatology. 2000;31:851-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 99] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 9. | Keeffe EB. Liver transplantation: current status and novel approaches to liver replacement. Gastroenterology. 2001;120:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 152] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 10. | Polimeno L, Margiotta M, Marangi L, Lisowsky T, Azzarone A, Ierardi E, Frassanito MA, Francavilla R, Francavilla A. Molecular mechanisms of augmenter of liver regeneration as immunoregulator: its effect on interferon-gamma expression in rat liver. Dig Liver Dis. 2000;32:217-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Yang L, Zhang YD, Liu XD. Construction of expression vector of GST-hALR fusion protein. Zhongguo Xiandai Yixue Zazhi. 2002;12:1-2. |
| 12. | Fan LQ, Zhang YD, Zhou SB. Experimental study of im-mortalized rat hepatocytes modified by enhanced green fluorescent protein gene. Zhongguo Xiandai Yixue Zazhi. 2002;12:7-9. |
| 13. | Lupp A, Danz M, Müller D, Klinger W. Expression and inducibility of cytochrome P450 isoforms in 1-year-old intrasplenic liver cell transplants in rats. Transpl Int. 2002;15:96-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 14. | Benoist S, Sarkis R, Barbu V, Honiger J, Baudrimont M, Lakehal F, Becquemont L, Delelo R, Housset C, Balladur P. Survival and functions of encapsulated porcine hepatocytes after allotransplantation or xenotransplantation without immunosuppression. Surgery. 2001;129:606-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Terada S, Matsuura K, Enosawa S, Miki M, Hoshika A, Suzuki S, Sakuragawa N. Inducing proliferation of human amniotic epithelial (HAE) cells for cell therapy. Cell Transplant. 2000;9:701-704. [PubMed] |
| 16. | Yu HF, Liu Q. [Expression of rat augmenter of liver regeneration in pichia pastoris and evaluation of its bioactivity in vitro]. Zhonghua Ganzangbing Zazhi. 2003;11:421-423. [PubMed] |
| 17. | Pan Y, He GQ, Li RB, Yi XR, Kong XP. [Modification of recombinant human augmenter of liver regeneration with urea studied by maldi-tof mass spectrometry]. Shengwu Huaxue Yu Shengwu Wuli Xuebao (Shanghai). 2003;35:355-359. [PubMed] |
| 18. | Gandhi CR, Kuddus R, Subbotin VM, Prelich J, Murase N, Rao AS, Nalesnik MA, Watkins SC, DeLeo A, Trucco M. A fresh look at augmenter of liver regeneration in rats. Hepatology. 1999;29:1435-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 19. | Wang A, Yang X, Wang W, Zuo F, Wang Q, He F. [Effect of recombinant human augmenter of liver regeneration on gene expression of tissue inhibitor of metalloproteinase-1 in rat with experimental liver fibrosis]. Zhonghua Yixue Zazhi. 2002;82:610-612. [PubMed] |
| 20. | Wang AM, Yang XM, Guo RF. Recombinant hALR to treat-ment experimental liver fibrotic rats. Zhongguo Ganbing Zazhi. 1999;7:243-246. |