Published online Jul 14, 2006. doi: 10.3748/wjg.v12.i26.4195
Revised: February 15, 2006
Accepted: February 28, 2006
Published online: July 14, 2006
AIM: To investigate human telomerase reverse transcriptase (hTERT) mRNA in the serum of cholangiocarcinoma patients.
METHODS: The serum of thirty three cholangiocarcinoma patients, forty one benign biliary tract disease patients and ten healthy volunteers were collected and analyzed for the expression of hTERT mRNA by real-time reverse transcriptase-polymerase chain reaction (RT-PCR). We then examined the correlation between values of serum hTERT mRNA and the pathological staging of cholangiocarcinoma.
RESULTS: hTERT mRNA was detected in 28 of 33 (84.85%) of serum obtained from cholangiocarcinoma patients and 9 of 41 (21.9%) of serum obtained from benign biliary tract disease patients. hTERT mRNA was not detected in any serum obtained from healthy volunteers. on the other hand the common tumor marker, CA19-9 was detected in 20 of 33 (60.6%) of serum obtained from cholangiocarcinoma patients and 8 of 41 (19.5%) of serum obtained from benign biliary tract disease patients. However, no correlation was found between the present of serum hTERT mRNA and tumor staging.
CONCLUSION: These results indicate that the detection of circulating hTERT mRNA was identified in almost all cholangiocarcinoma patients. It offers a novel tumor marker, which can be used as a complementary study for diagnosis of cholangiocarcinoma.
- Citation: Leelawat K, Leelawat S, Ratanachu-Ek T, Trubwongchareon S, Wannaprasert J, Tripongkaruna S, Chantawibul S, Tepaksorn P. Circulating hTERT mRNA as a tumor marker in cholangiocarcinoma patients. World J Gastroenterol 2006; 12(26): 4195-4198
- URL: https://www.wjgnet.com/1007-9327/full/v12/i26/4195.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i26.4195
Cholangiocarcinoma is the cancer arising from cholangiocyte, the epithelial cells lining the intrahepatic and extrahepatic bile ducts. It is one of the most common liver cancers in the population of Northeast Thailand and responsible for approximately one in five cancer-related deaths among Thai patients[1]. Three-year survival rates of 40%-60% have been reported only in a few number of patients resected for cure[2]. Diagnosis of cholangiocarcinoma is often difficult. It requires multiple complementary studies including evaluation of clinical symptoms, imaging, and tumor markers. Tissue biopsy and cytology have poor sensitivity and are positive only in about 30% of cases of cholangiocarcinoma. Recently, the percentages of positive serum obtained from the common marker (CA19-9) are only less than 70%[3]. In addition, CA19-9 can be elevated in cholestasis in the absence of malignancy, and following liver injury. Thus, their accuracy for the diagnosis of cholangiocarcinoma is limited. It is necessary to find novel markers to use in diagnosis and treatment.
The human telomerase, which composed of two subunits including telomerase RNA template (hTR) and telomerase transcriptase protein (hTERT), functions as a reverse transcriptase enzyme in the process of telomere synthesis[4]. Telomerase activity was detected in 85%-100% of cancer patients whereas normal somatic cells have low or undetectable[4,5]. In addition, previous results demonstrated that circulating tumor-related RNA including telomerase is frequently found in the plasma and serum of cancer patients[6-8].
Consequently, telomerase activity is possibly used as a common molecular tumor marker in the serum. Previous studies also found a good correlation between the telomerase activity and the expression of hTERT subunit. The aim of this study is to test the usefulness of hTERT mRNA detection in the serum of cholangiocarcinoma patients by using real-time reverse transcriptase polymerase chain reaction.
The human cholangiocarcinoma cell line HuCCA1 (kindly provided by Prof. Sirisinha, Department of Microbiology, Mahidol University) and RMCCA1 (established from Department of Surgery, Rajavithi Hospital) were grown in Ham’s F12 medium supplemented with 100 mL/L fetal bovine serum at 37°C in a 5% (50 mL/L) CO2 humidified atmosphere.
Thirty-nine informed and consenting patients undergoing surgery for cholangiocarcinoma at the Rajavithi Hospital, Thailand, between July 2003 and April 2006 were included in this study. Tumor samples were collected at the time of surgery and histopathologically characterized to confirm the diagnosis. Pathological data, including tumor staging was also collected. Fifty patients undergoing surgery for benign biliary tract disease were included in this study. Ten normal subjects were studied as negative controls.
Sample Collection
Blood was collected prior to surgery in plain tubes for serum sampling. After clotting, tubes were centrifuged at 1000 r/min for 15 min at room temperature, and serum was collected. This was followed by a second 15-min centrifugation at 1000 r/min to remove cellular debris. Serum samples were aliquoted and stored at -70°C until use. Serum CA19-9 level was measured in Clinical Laboratory of Rajavithi Hospital. The cut-off level was 100 IU/mL.
RNA Extraction
RNA from cell lines and serum was extracted using a commercially available kit (High Pure RNA Kit; Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer’s instructions. Only fresh or once-freeze thawed serum was used.
Quantitative detection of hTERT mRNA was performed with the TeloTAGGG hTERT Quantification Kit (Roche Diagnostics GmbH, Mannheim, Germany), using the LightCycler system (Roche Diagnostics, Mannheim, Germany) for real-time PCR according to the manufacturer’s instructions. For the reaction mixtures, 2 μL of hTERT reaction mix, 0.1 μL of reverse transcriptase, 2 μL of hTERT or PBGD mix, 13.9 μL of H2O and 2 μL of standard RNA template or RNA from serum samples was prepared. The reaction conditions were reverse transcription at 60°C for 10 min, followed by initial denaturation at 95°C for 30 s and 40 cycles of denaturation at 95°C for 0.5 s, annealing at 60°C for 10 s, and extension at 72°C for 10 s, respectively. The standard curve was established by determination of the five standards hTERT mRNA provided by the kit. The samples were normalized on the basis of the content of PBGD. Serum samples with more than 150 copies of PBGD suggesting an appropriate quality of RNA were used for the analysis of telomerase. Serum samples in which hTERT mRNA were detected were assigned to the hTERT-positive group.
Values were expressed as mean ± SD. Mean values were measured by the Student’s t test. Correlations between serum hTERT mRNA and stage of cholangiocarcinoma were assessed using the chi-square test (χ2). Sensitivity, specificity, positive predictive value, and negative predictive value were measured. P < 0.05 was considered as statistically significant.
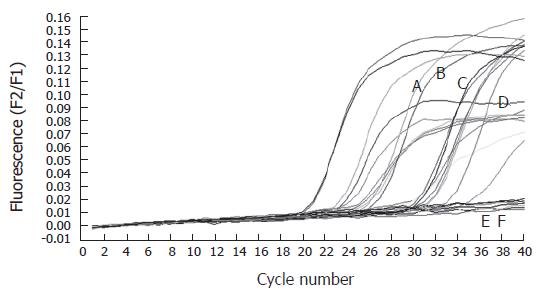
The expression of hTERT mRNA in two cholangiocarcinoma cell lines (HuCCA1 and RMCCA1) was investigated. Quantitative real-time RT-PCR demonstrated definite expression of hTERT mRNA in both cholangiocarcinoma cell lines (Figure 1). This evidence confirmed the existence of telomerase activity in cholangiocarcinoma. Therefore, we attempted to detect the circulating hTERT mRNA in the serum of cholangiocarcinoma patients.
Thirty-nine patients who were confirmed diagnosis as cholangiocarcinoma, fifty benign biliary tract disease patients and ten healthy volunteers were included in this study. Their serum was collected and extracted for total RNA. Only RNA samples, that could be detected by the expression of porphobilinogen deaminase (PBGD) as a housekeeping gene were included in this study. The thirty-three serum samples from cholangiocarcinoma patients, fortyone serum samples from benign biliary tract disease patients and ten serum samples from healthy volunteers detected for PBGD were assayed for the expression of hTERT mRNA (Figure 1). The patients’ demographic data was demonstrated in Table 1. There were no differences in sex, age, serum SGOT and serum SGPT between benign and cancer patients. However, total bilirubin and alkaline phosphatase were significantly high in cancer groups.
| Benign biliary tract disease | Cholangio-carcinoma | Healthyvolunteers | |
| Sex (Male:Female) | 21:20 | 19:14 | 6:4 |
| Age (year), (median) | 53.68 (22-82) | 56.40 (35-85) | 49.50 (26-60) |
| SGOT (IU/dL) | 105.32 ± 52.26 | 112.52 ± 44.54 | 30.20 ± 12.20 |
| SGPT (IU/dL) | 96.04 ± 45.42 | 73.41 ± 44.30 | 28.50 ± 9.22 |
| Total Bilirubin (mg/dL)a | 3.8 ± 2.21 | 12.8 ± 5.24 | 1.05 ± 0.25 |
| Alkaline Phosphatase (U/dL)a | 309 ± 67.53 | 550 ± 24.44 | 98 ± 10.60 |
Serum hTERT mRNA was recognized in 28 of 33 cholangiocarcinoma patients (84.85%) and 9 of 41 benign biliary tract disease patients (21.9%). However, serum hTERT mRNA was not detected in any healthy volunteers. The efficiency of serum hTERT was compared with the serum CA19-9 as shown in Table 2. Serum hTERT was higher in sensitivity for detection of cholangiocarcinoma than serum CA19-9.
| Serum hTERT (%) | Serum CA19-9 (%) | |
| Sensitivity | 84.85 | 60.6 |
| Specificity | 78.05 | 80.49 |
| Positive predictive value | 75.68 | 71.43 |
| Negative predictive value | 86.49 | 71.74 |
| False Negative | 13.51 | 28.26 |
| False Positive | 24.32 | 28.6 |
We also evaluated for the association between serum hTERT mRNA and the histopathological staging of cholangiocarcinoma in the surgical specimens resected from these patients. The result showed that serum hTERT mRNA could be detected in all stages of cholangiocarcinoma patients. However, it did not correlate with the staging of cholangiocarcinoma (Table 3).
| Tumor stages | Serum hTERT | mRNA |
| positive | negative | |
| Stage I | 2 | 1 |
| Stage II | 17 | 3 |
| Stage III | 9 | 1 |
Eukaryotic chromosomal ends consist of repeating DNA sequences (TTAAGG) termed telomeres. An enzyme that adds telomeric repeats onto chromosomal ends is telomerase. This enzyme is composed of two subunits; hTR and hTERT[4]. Accordingly, both hTR and hTERT are necessary for telomerase activity, yet the catalytic activity of the enzyme is generally regulated through the presence and activity of hTERT. Therefore, the detection of hTERT mRNA is a guarantee for the present of telomerase activity[8]. In most somatic cells in which telomerase activity is undetectable, telomeric sequences are lost with each cell division because of the end-replication problem. Unlike healthy cells, most malignant human cells are capable of escaping senescence and sustaining infinite proliferation through the activation of telomerase to stabilize their telomere length[4,8-12].
Traditionally, telomerase activity has been assessed based on a biochemical primer extension assay, the inefficiency and low sensitivity of which, together with the low amounts of telomerase activity, greatly limit the application of the assay in primary human tumors[9]. Therefore, the detection of hTERT by real-time RT-PCR was used in this study. This assay determined the expression of telomerase by measuring the amount of the mRNA encoding its catalytic subunit hTERT. The relative telomerase expression levels were determined by comparing them to the expression levels of the housekeeping gene porphobilinogen deaminase (PBGD). hTERT-encoding mRNA from the serum was reverse transcribed and 198 bp fragments of the generated cDNA was amplified with specific primers in a one step RT-PCR reaction. The amplicon was detected by fluorescent emission using a specific pair of hybridization probes. In a separate RT-PCR, mRNA encoding for porphobilinogen deaminase (PBGD) was processed. The reaction product served as both a control for RT-PCR performance and as a reference for relative quantification.
Our study showed that hTERT mRNA was detected in almost all of the cholangiocarcinoma patients (84.85% of cases). However, in benign biliary tract disease patients, hTERT mRNA was also detectable (21.9% of cases). According to the previous report, telomerase activity has been reported in normal lymphocytes[8-12]. This result suggested the contamination of lymphocytes in the serum specimens. Comparison with the common tumor marker, CA19-9 was detected in only 60.60% of cases. This data suggested that hTERT mRNA should be a candidate tumor marker in cancer patients.
hTERT mRNA is not specifically detected in serum of cholangiocarcinoma patients but it was also significantly found in several types of cancers such as breast cancer, malignant melanoma and thyroid cancer[8]. Certainly, we have detected hTERT mRNA in the serum of five patients with hepatocellular carcinoma and three patients with pancreatic cancers (data not shown). This indicates that the detection of circulating extracellular tumor-derived mRNA is not confined to any one cancer type, but may actually be a relatively ubiquitous finding across a broad range of cancers. However, no relationship was found between the expression of hTERT mRNA and clinicopathological findings. According with previous study, the presence of hTERT was unrelated to tumor size, tumor grade or the presence of nodal metastasis[12]. These results suggest that the detection circulating hTERT mRNA does not predict prognosis in cholangiocarcinoma.
In conclusion, hTERT might serve as a tumor marker, which early identified circulating specific RNA originating from tumor cells. However, further examinations using more cholangiocarcinoma cases are necessary to evaluate the usefulness of this marker.
S- Editor Wang J L- Editor Olaleve SB E- Editor Liu Y
| 1. | 1 Sriplung H, Sontipong S, Martin N, Wiangnon S, Vootiprux V, Cheirsilpa A, Kanchanabat C, Khuhaprema T. Cancer incidence in Thailand, 1995-1997. Asian Pac J Cancer Prev. 2005;6:276-281. [PubMed] |
| 2. | Chamberlain RS, Blumgart LH. Hilar cholangiocarcinoma: a review and commentary. Ann Surg Oncol. 2000;7:55-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 92] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Olnes MJ, Erlich R. A review and update on cholangiocarcinoma. Oncology. 2004;66:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 187] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Bachand F, Triki I, Autexier C. Human telomerase RNA-protein interactions. Nucleic Acids Res. 2001;29:3385-3393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Yan P, Coindre JM, Benhattar J, Bosman FT, Guillou L. Telomerase activity and human telomerase reverse transcriptase mRNA expression in soft tissue tumors: correlation with grade, histology, and proliferative activity. Cancer Res. 1999;59:3166-3170. [PubMed] |
| 6. | Johnson PJ, Lo YM. Plasma nucleic acids in the diagnosis and management of malignant disease. Clin Chem. 2002;48:1186-1193. [PubMed] |
| 7. | Fleischhacker M, Beinert T, Ermitsch M, Seferi D, Possinger K, Engelmann C, Jandrig B. Detection of amplifiable messenger RNA in the serum of patients with lung cancer. Ann N Y Acad Sci. 2001;945:179-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Novakovic S, Hocevar M, Zgajnar J, Besic N, Stegel V. Detection of telomerase RNA in the plasma of patients with breast cancer, malignant melanoma or thyroid cancer. Oncol Rep. 2004;11:245-252. [PubMed] |
| 9. | Mabruk MJ, O'Flatharta C. Telomerase: is it the future diagnostic and prognostic tool in human cancer. Expert Rev Mol Diagn. 2005;5:907-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy HE, Meyer P, Stroun M. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823-3826. [PubMed] |
| 11. | Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1114] [Cited by in RCA: 1203] [Article Influence: 60.2] [Reference Citation Analysis (1)] |
| 12. | Li YR, Wu JM, Wang L, Huang X, Shi J, Hu LH. Human telomerase reverse transcriptase expression and its clinical significance in laryngeal squamous cell carcinoma. Acta Otolaryngol. 2005;125:409-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |